This page is dedicated the bGWAS R-package. The method has been published in Bioinformatics (N. Mounier & Z. Kutalik, 2020) and the R-package is available here.
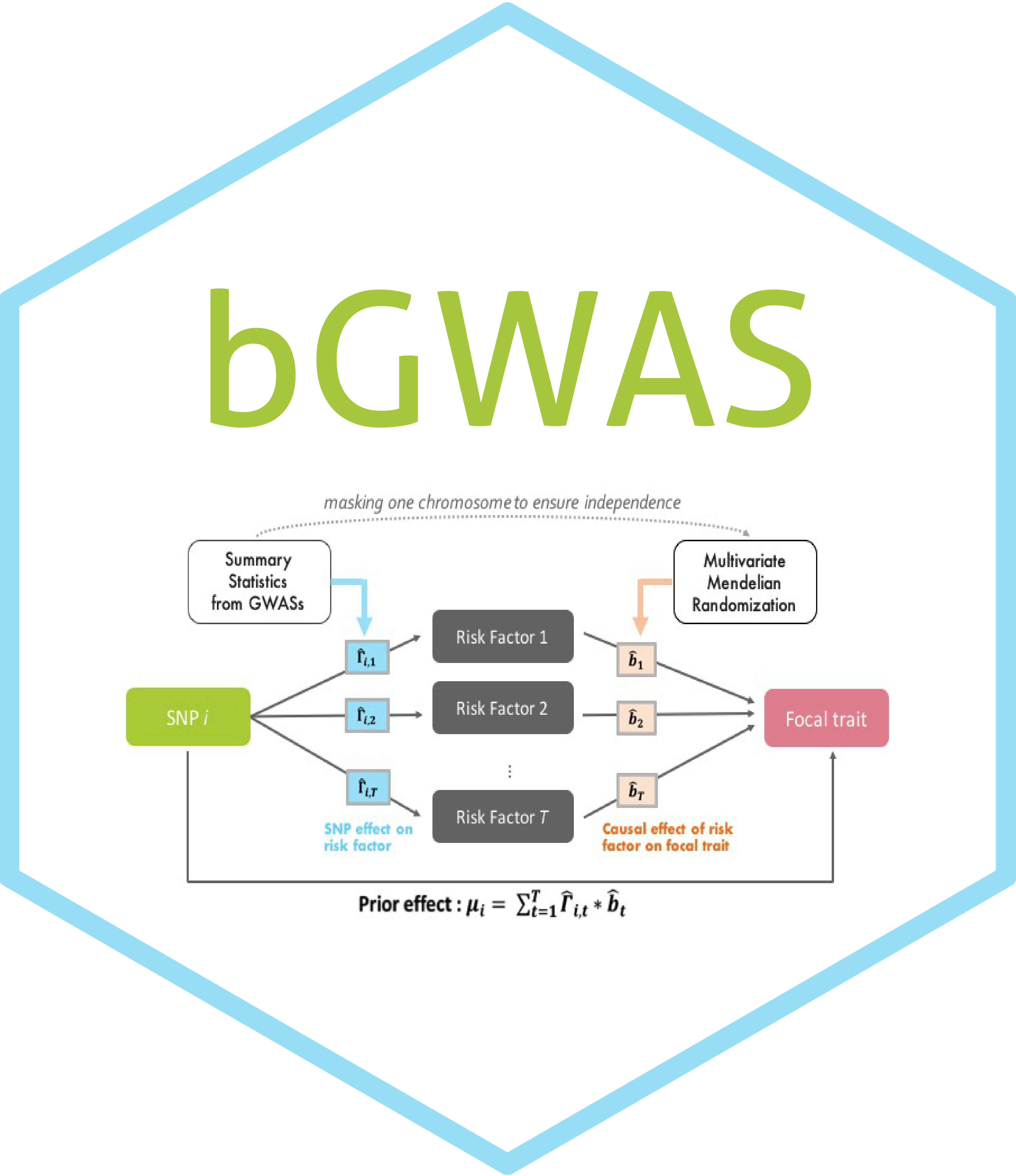
bGWAS is an R-package to perform a Bayesian GWAS (Genome Wide Association Study), using summary statistics from a conventional GWAS as input. The aim of the approach is to increase power by leveraging information from related traits and by comparing the observed Z-scores from the focal phenotype (provided as input) to prior effects. These prior effects are directly estimated from publicly available GWASs (hereinafter referred to as “prior GWASs” or “risk factors”). Only prior GWASs having a significant causal effect on the focal phenotype, identified using a multivariable Mendelian Randomization (MR) approach, are used to calculate the prior effects. Causal effects are estimated masking the focal chromosome to ensure independence, and the prior effects are estimated as described in the figure below.
Observed and prior effects are compared using Bayes Factors (BFs). Significance is assessed by calculating the probability of observing a value larger than the observed BF (P-value) given the prior distribution. This is done by decomposing the analytical form of the BFs and using an approximation for most BFs to make the computation faster. Prior, posterior and direct effects, alongside BFs and p-values are returned. Note that prior, posterior and direct effects are estimated on the Z-score scale, but are automatically rescaled to beta scale if possible.

