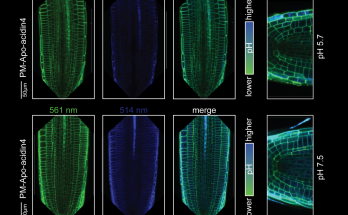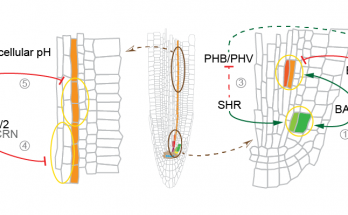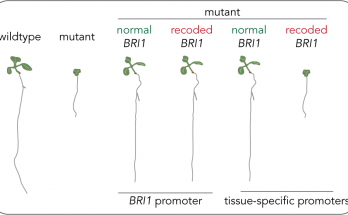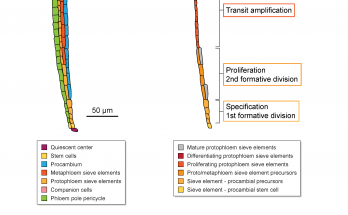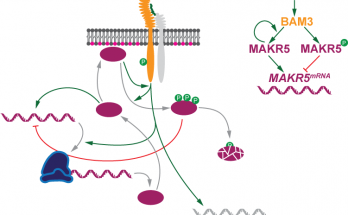
Something’s incoherent here
…and it’s a feedforward loop! Qian’s discovery of a nontranscriptional incoherent feedforward loop in a receptor kinase signaling pathway has now been published in the Proceedings of the National Academy …
Something’s incoherent here Read More