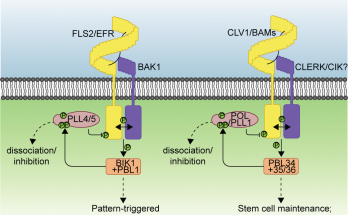
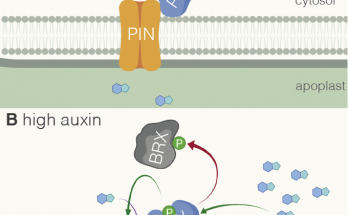
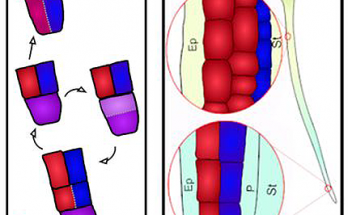
The grey zone between natural and artificial exposed
Who would have thought BRX mutants and transgenics will make it to the museum one day? An intriguing exposition at the Natural History Museum in Sion explores the question where …
The grey zone between natural and artificial exposed Read More