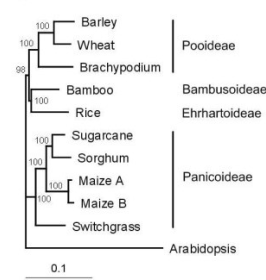
The dicotyeyledon model species Arabidopsis thaliana is a great system to work with and will likely stay unmatched for analyses at the cellular to subcellular level for a long time to come. Yet at least genetic verification of findings obtained from Arabidopsis is possible in tractable monocotyledon systems, such as Brachypodium distachyon. Brachypodium is a close relative of major European crops: Brachypodium belongs to the Brachypodieae, which is a basal tribe within the Pooideae, the monocotyledon subfamily that includes the major European crops barley, rye and wheat. Brachypodium is therefore phylogenetically much closer to these crops than any other monocotyledon model, including rice or corn. Moreover, unlike these domesticated species, Brachypodium is a wild plant. Nevertheless, the phylogenetic proximity of Brachypodium to barley, rye and wheat is also observed at the genomic level, as exemplified by the high degree of homology and overlap between the gene sets of Brachypodium and wheat (protein sequence identity of the 45,318 best bidirectional hits between Brachypodium and wheat is 76.3% on average, with a median of 80.3%), suggesting that allelic variants of interest that have been functionally defined in Brachypodium can be exploited in those crops, for instance through smart breeding. While gene functions and hypotheses obtained from Arabidopsis research are often directly applicable to monocotyledons such as Brachypodium in general, in detail we often observe interesting differences, such as alternative genetic wiring that sometimes results in unexpected outcomes. Pertinent publications on this topic include the following: