“I think I need a bigger box.”
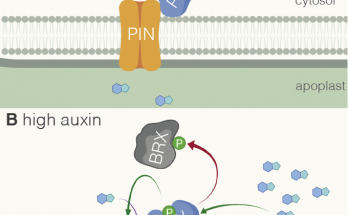
…for my bigger seeds. Sam’s paper, building on his keen sense of observation, is now out in Development: We found that heterologous expression of certain BRX variants boosts seed size …
“I think I need a bigger box.” Read More