Overview
Being early adopters of the Pacific Biosciences sequencing technology, we now have more than 8 years’ experience with long read sequencing. We currently run the latest Pacific Biosciences Sequel II system.

Main applications are:
• De novo genome sequencing (any type, from small bacterial genomes to large, complex animal or plant genomes)
• Nucleotide and structural variant analysis
• DNA base modification analysis (epigenetics)
• Full length RNA isoform sequencing (including single cell analysis)
• Long amplicon sequencing (1 kb to 10 kb, Sanger-like quality)
PacBio SMRT sequencing
Pacific Biosciences libraries molecules are circular
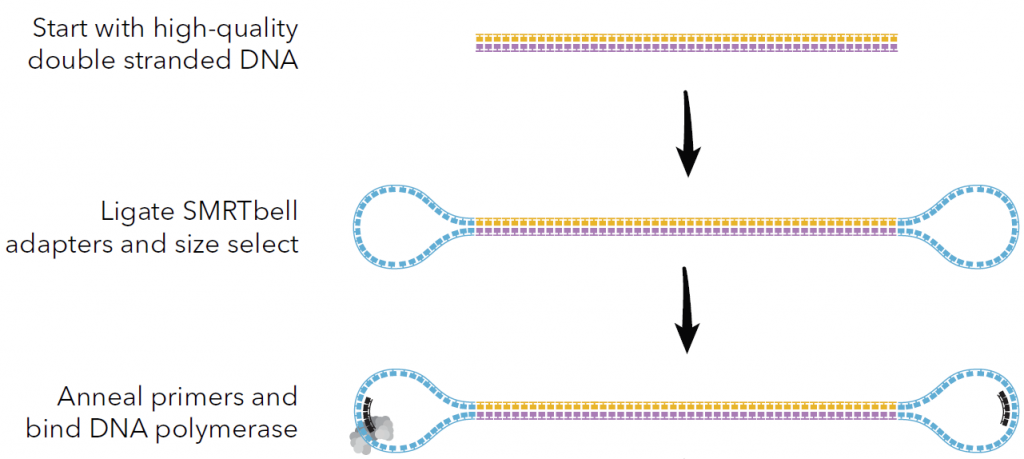
Source: Pacific Biosciences
They are loaded in a SMRT cell for Single Molecule Real Time sequencing. Sequencing movies can last for up to 30 hours.
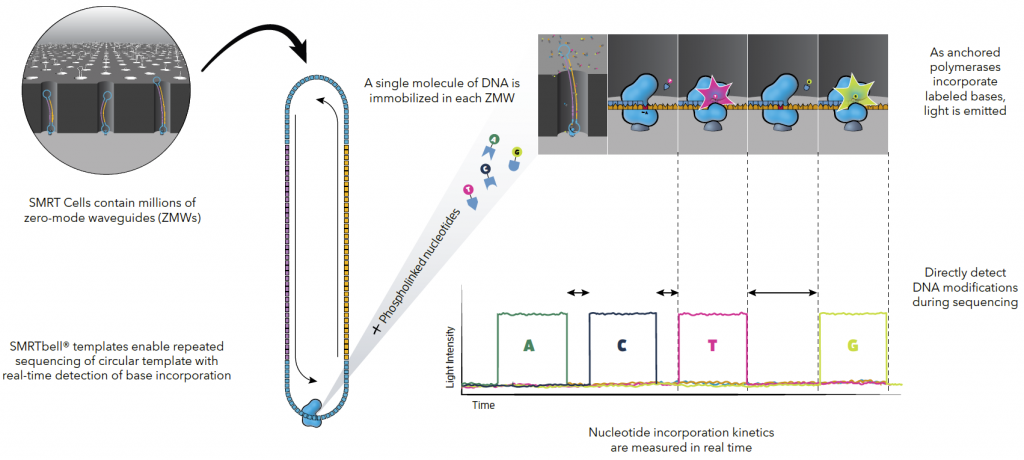
Source: Pacific Biosciences
A SMRT cell is 8M wells in total. Loading must be fine-tuned to ensure maximum occupancy without getting multiple Library/Polymerase complexes per well. Depending on the library type, the output is generally 1M to 6M reads per run.
The actual yield in Gb varies depending on the library quality and can typically reach more than 500 Gb with optimal conditions.
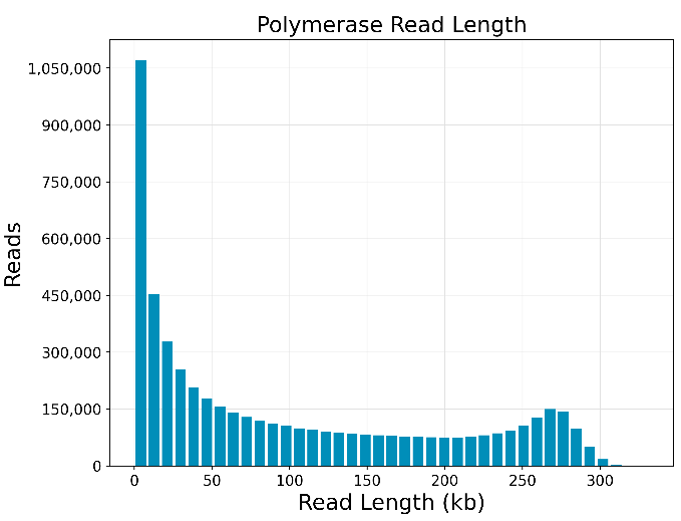
In house example of a 500 Gb raw read output run: Read length distribution
HiFi reads
Because Pacific Biosciences library molecules are circular, the same DNA insert molecule, if not too long, can be read many times over the course of a sequencing run. All the raw reads coming from the same single insert can be aligned and corrected, generating a high quality read called CCS (for Circular Consensus Sequence, see figure below) or HiFi read.
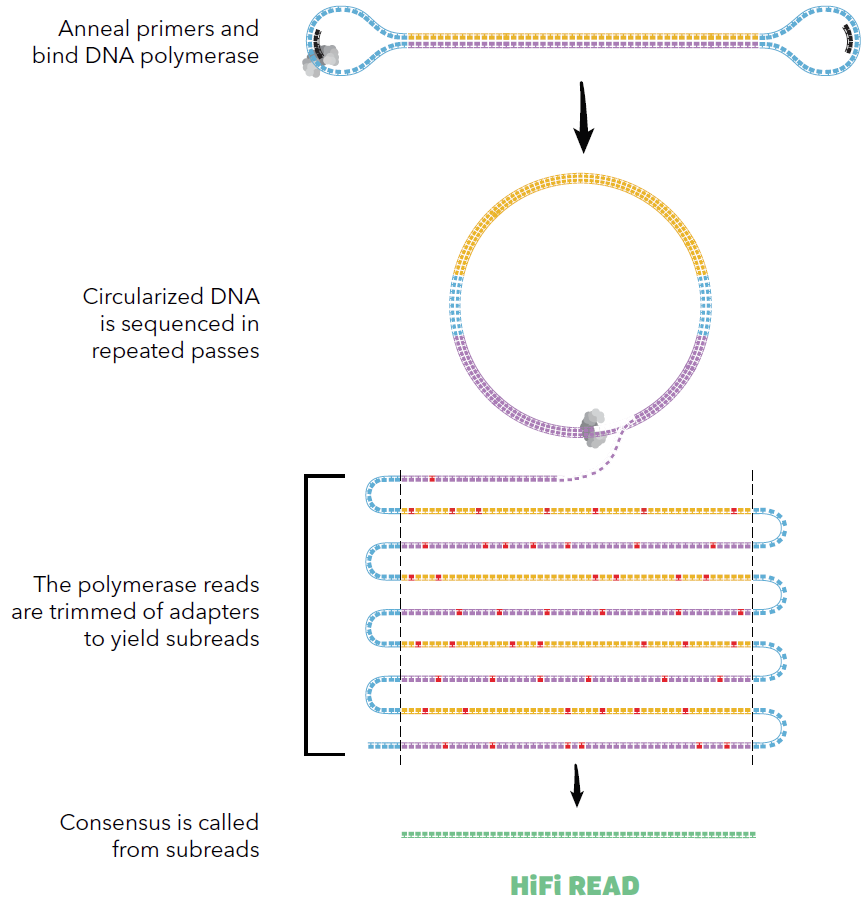
Source: Pacific Biosciences
Typical CCS read metrics (read length and quality) are shown below.
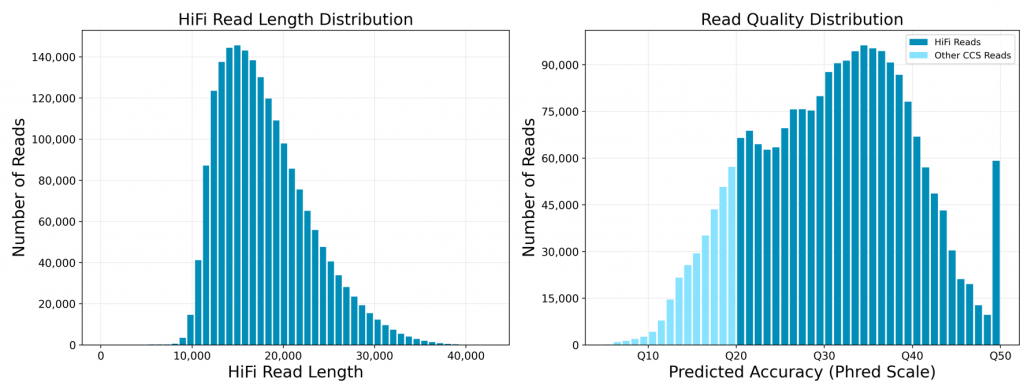
HiFi reads are not ultra-long but, thanks to their ultra-high accuracy and uniform coverage over the genome (no GC bias), they are ideal for many applications such as phased assembly and variant analysis. No short sequencing read based error correction is required.