Overview
Short read sequencing is operated with Illumina or Element Biosciences technologies.

The main applications are:
- Transcriptome analysis (total RNA, messenger RNA, small RNA)
- Whole genome sequencing
- Whole exome sequencing
- Targeted sequencing (e.g. ChIP-Seq, 16S, PCR amplicons, capture)
Single end or paired end sequencing
There are two possible protocols for short read sequencing workflows: Single End (SE, also called SR for Single Read) and Paired End (PE) sequencing.
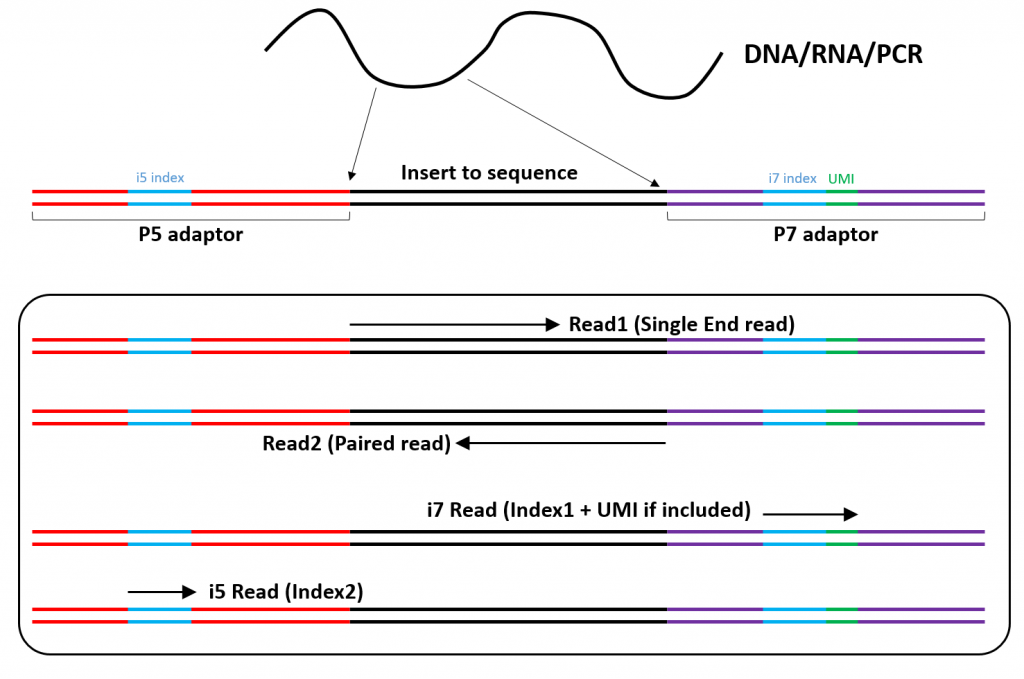
- With Single Read sequencing, the library insert is read from the P5 end only (see figure below). With Paired End sequencing, a second read is generated from the other end of the insert (P7 end).
- Paired End sequencing “doubles” the sequencing coverage for a given number of sequencing lanes. It requires an insert long enough to minimize the overlap of the two corresponding reads. It is used for whole genome or whole exome sequencing, and in specific cases of transcriptome sequencing.
- Single End sequencing is the method of choice for RNA-sequencing, if the aim is differential gene expression analysis.
Indexing options
The output of one sequencing lane is generally too high for a single library. Multiple libraries are therefore pooled and sequenced together. Each sequencing read is subsequently re-attributed to its sample of origin thanks to a barcode (also called “index”) incorporated in library adapter (see figure below).
There are several indexing strategies:
- Single indexing (i7 index only). It is available for any library type. It is however not recommended for sequencing on Illumina patterned flow cells (NovaSeq, HiSeq4000, NextSeq2000) due to index swapping issues (wrong index assigned to some sequencing reads, see Illumina website). This is not an issue for sequencing with Element Biosciences.
- Combinatorial Dual Indexing (i7 and i5 indexing). It increases the number of barcodes available but suffers from the same swapping weakness as single indexing.
- Unique Dual Indexing (UDI). It is a dual indexing strategy in which each i7 and i5 barcode is unique. It prevents index swapping issues and is considered as good practice for Illumina sequencing.
- Unique Molecular Identifier (UMI) can optionally be used for ultimate sequencing reads deduplication.