The GTF facility provides bioinformatic services for (1) primary analysis and quality control of genomic data, (2) data storage (new politics soon, according UNIL rules) and transfer, (3) data processing, and (4) statistical analysis.
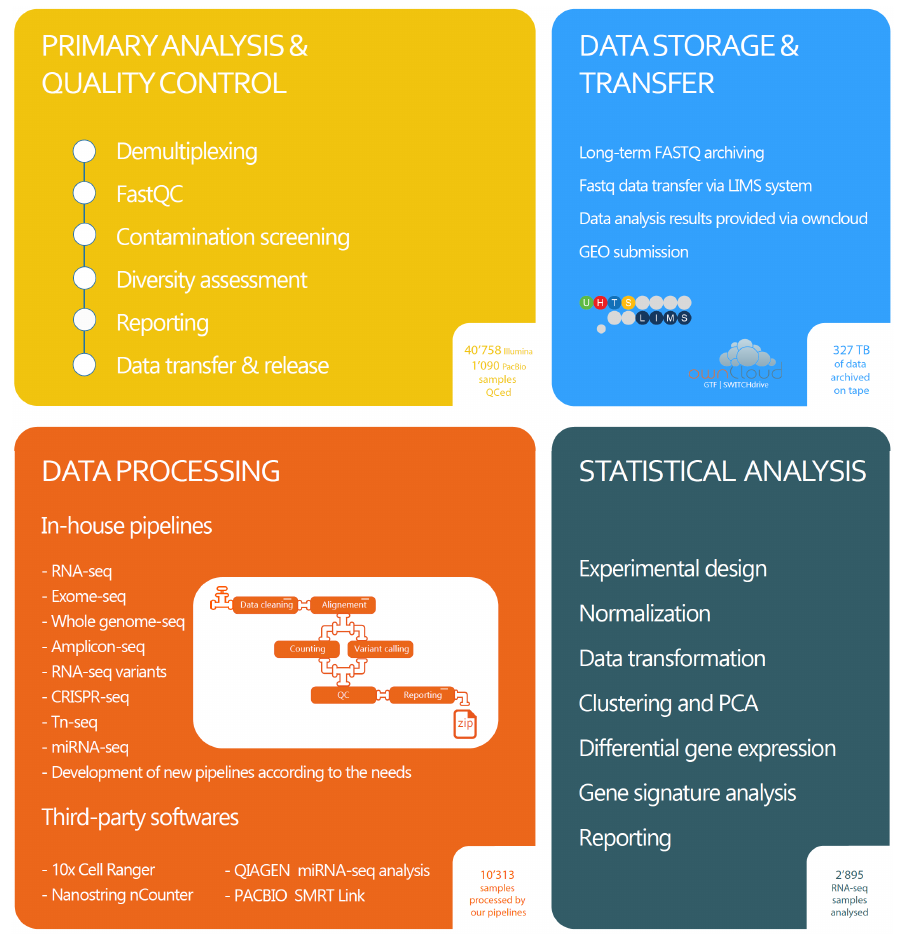
Primary analysis and quality control of genomic data
Raw sequencing data are converted into standard ‘FASTQ’ files. A set of automated QC procedures are applied in order to identify possible technical biases or issues during sample preparation, library preparation, or sequencing. This includes for instance the search for unexpected index, and the mapping over multiple reference genomes.Data storage and transfer
FASTQ files are archived for long term storage (new politics soon, according UNIL rules). They can be retrieve upon request and are accessed through our LIMS system. For project analyzed by the GTF, we provide also support for GEO submission.
Data processing
The GTF provides application-specific quality controls and data processing pipelines. Data processing pipelines include sequence read cleaning, mapping, counting and/or variant calling and reporting. Custom pipelines may be developed upon demand.
Experimental design and statistical analysis
The GTF provides assistance and counseling in designing mRNA-seq, miRNA-seq and total RNA-seq experiments. Even in the most carefully performed investigations, poor experimental planning and design can result in the generation of useless and/or un-interpretable data. This can very often be avoided by including statistical considerations during the experimental design stage. Users are strongly encouraged to make use of the bioinformatic and statistical consultation services provided by the GTF when designing their RNA-seq projects – BEFORE ANY – steps of the experiment have been carried out.
For RNA-seq experiments, statistical analysis can be conducted up to the point of lists of differentially expressed genes and isoforms. Data visualization includes PCA, Venn diagrams, Volcano plots and heatmaps. Assistance in high-level statistical analysis can be discussed in a case-by-case manner (e.g for gene ontology or pathway analysis).

