3 Short Reads sequencers are currently operated by GTF: NovaSeq and MiSeq from Illumina, and Aviti from Element Biosciences
Sequencing prices are available on our LIMS
NovaSeq 6000
This is one of the latest sequencer from Illumina. It offers high output for a very competitive price for large flow cells (S4, and to a lesser extent S2).
4 types of flow cells with different outputs are available.
All flow cells are patterned and ideally require libraries with Unique Dual Indexing.
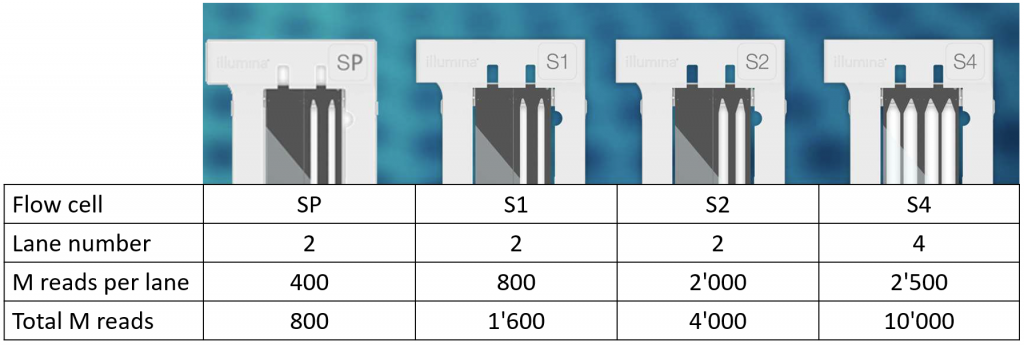
Flow cells can be used for 100 cycles, 200 cycles, or 300 cycles sequencing runs
These cycles can be split and used either for single end (SR or SE) or paired end runs (PE)
Please discuss actual limitations with GTF staff
Standard settings proposed by GTF are SR 100 cycles / PE 150 cycles / PE 28+90 cycles (10X single cell settings).
The entire flow cell (2 or 4 lanes) must be processed with the same cycling conditions. Requests not matching these standard settings may therefore wait longer (to fill the flow cell) before being processed.
WARNING: Data generated on the NovaSeq may not directly compare to legacy HiSeq data
• NovaSeq is a 2-colors system while HiSeq were 4 colors
• Novaseq goups Q-scores into 4 bins while HiSeq used > 70 Q-scores (see details)
↠ This change is mostly neutral for gene expression analysis (e.g. RNAseq, bulk or single cell)
↠ It may have a non negligible impact on variant analysis
↠ Analysis pipeline codes may require updating
Aviti
Element Biosciences sequencer Aviti is an alternative to Illumina sequencing for moderate throughput short read projects.
Its main characteristics are:
- Sequencing of 500M reads per lane (2 lanes available)
- Price equivalent to NovaSeq S4 flow cell (around half price of the smaller SP flow cell)
- Sequencing quality superior to Illumina (Q40 instead of Q30, less sensitivity to low diversity)
- Not subject to index swapping issue
This sequencer is fully compatible with standard Illumina libraries. An extra processing step is required but this is performed at no extra cost by the facility, and it does not involve any extra PCR step.
Please inquire for more technical details
MiSeq
MiSeq sequencing offers the highest flexibility but is limited by a relatively low throughput. It is therefore recommended for projects that do not require large number of reads, such as targeted sequencing or small genome sequencing. The “nano” option is an inexpensive option for evaluating library quality or pool equimolarity.
- 1M to 25M reads
- Single End or Paired End runs
- Custom read length from 50 nt to 300 nt
| MiSeq Kit | Readlength (Total nt) | Million reads* |
|---|---|---|
| v2 nano | 300 | 1 |
| v2 nano | 500 | 1 |
| v2 micro | 300 | 4 |
| v2 | 50 | 15 |
| v2 | 300 | 15 |
| v2 | 500 | 15 |
| v3 | 150 | 25 |
| v3 | 600 | 25 |
* Low diversity library sequencing (e.g. 16S or other amplicon based libraries) requires the presence of a stuffer library to artificially increase base content diversity. This reduces the sequencing throughput by 10% to 50%, depending on the spike in level
HiSeq 4000 (discontinued end of 2021)
- 300 M reads per lane for standard library types
- Single End or Paired End runs
- Standard sequencing read length is 150 n
Because of HiSeq4000 index swapping issues (wrong index assigned to some sequencing reads, for details see Illumina website), libraries sequenced on this machine require a unique dual indexing strategy (UDI).
Non-standard read length is available upon demand but must involve a request for an entire flow cell (8 lanes, 2’400M reads).
HiSeq 2500 (discontinued end of 2021)
- 200 M reads per lane for standard library types
- Single End or Paired End runs
- Standard read length is 125 nt for Single End and 100 nt for Paired End
Index swapping is not an issue for sequencing on HiSeq2500. It can be used with any library preparation protocol.
Due to the fact that the sequencing price is equal between HiSeq2500 and HiSeq4000, few requests are submitted for HiSeq2500, resulting in prolonged waiting times. We therefore propose this service either as a full flow cell run (8 lanes) or in a so-called Rapid Mode (see below).
Non-standard read length is available upon request but must involve a request for an entire flow cell (8 lanes).






