We offer several sequencing library preparation workflows for short read sequencing (Illumina) applications.
Generic sample quality and quantity guidelines for standard library preparation can be found HERE.
Libraries preparation includes fluorimetric method based quantification and size pattern quality control. When necessary, quantitative PCR and MiSeq sequencing runs can be proposed as optional quality control steps.
Most libraries are generated with unique dual indexing for compatibility with the HiSeq4000. We can optionally propose to include UMI (Unique Molecular Identifiers) for ultimate sequencing read deduplication.
Single cell libraries are discussed in another section.All library preparation and sequencing requests must be submitted through our LIMS system.
DNA library preparation protocols proposed by the GTF
- Standard DNA library preparation (involves DNA shearing and PCR amplification steps). Optimal input is 100 ng but the methods can go as low as 10ng.
- Tagmentation based library preparation for increased throughput. We use Nextera XT for small genomes (1 ng input) and Nextera FLEX for large genomes (optimal input 100 ng).
- PCR-free library preparation for more homogenous coverage in difficult to amplify regions (e.g. GC rich). This requires 1 ug DNA input and is not suitable for FFPE samples.
- Low input library preparation when DNA available is below 1 ng (e.g. ChIP-Seq experiments).
- Exome sequencing library preparation. Whole exome sequencing is available with a variety of library preparation and capture options, including Agilent SureSelect, IDT, and Twist Bioscience kits. Such capture protocols are not restricted to whole exome and can be used for small targeted regions (e.g. a single gene). For small targets, we can offer custom made probes (please, contact us for further information).
- Low Pass Sequencing (around 1X coverage or lower) can be a cost effective strategy for identifying variant within large cohorts. We use a highly multiplexed strategy (from seqWell) so that the library preparation cost is not prohibitive.
DNA processed upstream of the library preparation steps (e.g. bisulfite treatment, chromatin immunoprecipitation) is welcome for library preparation, but the GTF does not offer this upstream processing.
RNA library preparation protocols proposed by the GTF
- Standard messenger RNA library preparation (for polyadenylated RNA) for standard differential gene expression analysis. Optimal input is 500 ng but the methods can go as low as 10ng. RNA must be of high quality (RIN/RQN > 8.0, see this publication for further information).
- Total RNA library preparation for degraded RNA, or projects involving non polyadenylated RNA analysis. Inputs are the same as for mRNA libraries. Total RNA sequencing libraries are more expensive to prepare (since it includes a ribosomal RNA depletion step), and to sequence (not only mRNA so more complexity).
- Low input RNA library preparation when 10 ng RNA is not available. This can go virtually as low as single cell (few pg RNA). It works for mRNA or total RNA analysis. Low input protocols are available, either with direct library preparation, or with a cDNA amplification step. The latter option allows for quantitative PCR control of gene expression (by the user) before proceeding with the actual library preparation.
- Small RNA library preparation from miRNA profiling for instance. This optimally requires 100 ng of high quality RNA (RIN/RQN > 8.0) but works out of this specification range (please inquire). RNA must be extracted with a method compatible with small RNA purification.
Amplicon sequencing
Any PCR product can be sequenced on an Illumina platform provided it carries P5 and P7 sequences at its extremities. This is for instance used for 16S sequencing, CRISPR library screening, or sequencing of CRISPR edited sites.
There are 2 possible strategies (see figure below):
- The PCR primers carry P5 and P7 tails. This is the most straightforward option but multiplexing requires ordering multiple similar primers pairs with distinct barcodes.
- The PCR primers carry overhang sequences to which Illumina P5/P7 primers hybridize in a second round of PCR. This option is generally preferred since it relies on a single, tailed, gene specific primer pair while universal barcoded P5 / P7 primers are readily available from Illumina (up to 384-plex pooling). A 2-step protocol for 16S amplifications is available from Illumina
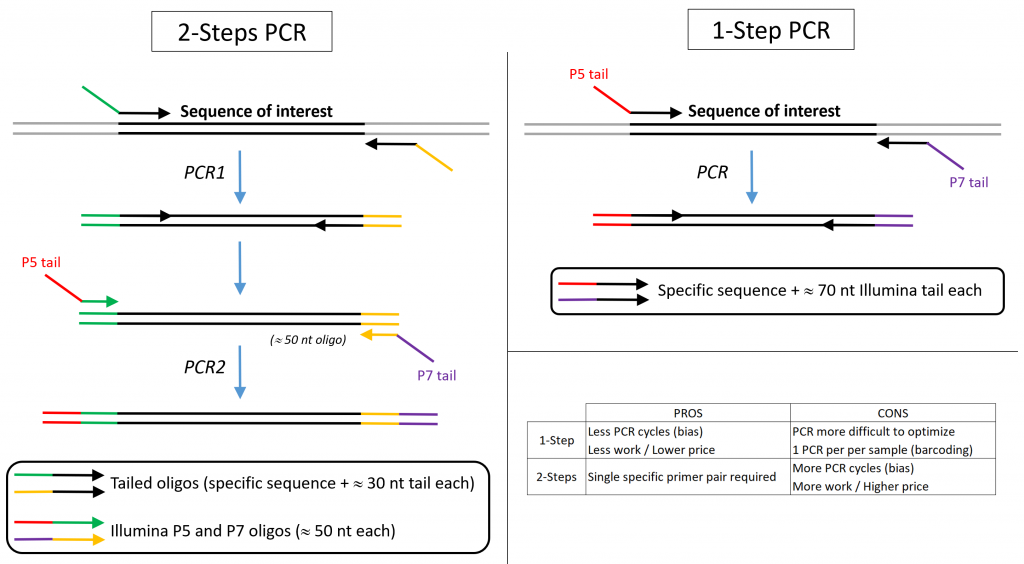

We offer the flexibility to start amplicon sequencing library preparation at any step depending on your needs. The first (target specific) PCR remains of user’s responsibility.
| Step # | Step |
|---|---|
| 1 | First PCR purification |
| 2 | First PCR quantification |
| 3 | Second PCR (indexing) |
| 4 | Second PCR purification |
| 5 | Individual PCR products QC |
| 6 | PCR products pooling |
Depending on your project, long read sequencing may be more informative and less expensive than a short read amplicon sequencing strategy (see full length bacterial 16S sequencing).

