Introduction
Sex is a stereotypical process where two haploid genomes come together to build a diploid, which then undergoes meiosis to half the ploidy. To ensure faithful succession of two ploidy states, eukaryotes specify distinct cell fates that restrict mating to haploid gametes and meiosis to diploid zygotes. How fertilization triggers a gamete-to-zygote fate switch is a fundamental question in developmental biology that we focus on.
We use fission yeast Schizosaccharomyces pombe as a model system with powerful genetics, genomics, cell biology and biochemical approaches. Fission yeast is a haploid organism that reproduces asexually, through mitotic divisions when nutrients are available (Video 1). However, when nitrogen is depleted from the environment, cells arrest the mitotic cycle and commit to sexual reproduction. Cells differentiation into P- and M-gametes by expressing mating-specific genes including those producing diffusible pheromones and their receptors. Partner gametes then communicate through pheromones to orient growth that brings them into physical contact. Two partners subsequently fuse to form a diploid zygote that rapidly initiates meiosis followed by production of four haploid spores that are resilient to environmental changes.
Parent-specific expression of genes
We found that the initiation of the diploid-specific processes relies on a complex between a homeodomain transcription factor Pi, which is expressed only in P-gametes, and a short peptide Mi, which is induced in M-gametes only (Figure 1). Prior to fertilization Pi is nuclear while Mi localizes to the cytosol. Though expressed, neither Pi nor Mi are active in their respective gametes since neither binds target DNA sequences individually. After fertilization however, Pi captures Mi and recruits it to the nucleus where the two form a complex that initiates zygote-specific transcription. The Pi-Mi complex provides a stunning example of transcriptional activation
in vivo, which we use to explore
how small peptides regulate transcription factors.
Since Pi is nuclear in the P-gamete, Mi is rapidly recruited to the P-nucleus and only later to the M-nucleus (Video 2). Consequently, the Pi-Mi complex induces transcription from the P-parental genome first. One target of the Pi-Mi complex is the zygotic inducer gene mei3, which we show is expressed more rapidly and robustly from the P-parental allele (Figure 2). Remarkably, this underlies a functional difference between two parental mei3 alleles: While deletion of the “slow” M-allele has little effect, ablation of the “fast” P-allele causes a delay in zygote specification such that zygotes do not arrest mating and instead fuse with additional partners (Video 3).
Our work was the first to show that a simple, two component transcription factor can generate asymmetry between parental alleles. Furthermore, it introduces fission yeast as a novel, powerful model system where experimental evolution is feasible to study parent-biased gene expression, which was previously considered to occur only in mammals and flowering plants. Our current work aims to identify the prevalence and roles of parent-specific allelic expression in fission yeast, as well as the mechanisms that generate it.
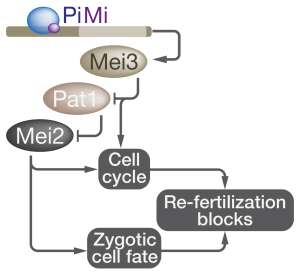
Re-fertilization blocks
After two haploid cells fuse, the newly formed diploid must avoid further mating or risk fusing with additional partners to form polyploids. While mechanisms that prevent consecutive fertilizations have been reported in animals and plants, our work provides the first insight into fungal blocks to re-fertilization. After gametes fuse, the Pi-Mi complex drives rapid induction of Mei3 to block mating. If Mei3 expression is delayed, zygotes are seen engaging and fusing with additional partners (Video 3). We find that Mei3 blocks re-fertilization in two ways (Figure 3): Firstly, Mei3 directly stimulates the zygotic exit from the G
1-phase of the cell cycle, which is the only stage when cells can mate. Secondly, Mei3 initiates a signaling cascade that results in activation of Mei2, an RNA-binding protein that block mating independently of the G
1-exit through mechanisms that remain unknown.
Our current aims is to reveal the molecular networks that block mating in response to Mei3 expression and downstream Mei2 activation . Understanding fungal re-fertilization blocks may, on one hand, identify mechanistic principles conserved between eukaryotic lineages, as increasingly evident for other aspects of sexual reproduction such as cell-cell fusion. On the other hand, re-fertilization blocks in a microbial context may have unexpected, adaptive roles, since ploidy alterations may increase fitness under certain conditions in both non-pathogenic and pathogenic fungi.
Zygotic cell fate
The lack of tools to study the short time zygotes spend in G1-phase has limited our understanding of zygotic specification prior to meiotic commitment (Video 3). For example, targets and molecular mechanisms of Mei2 regulation remain largely unknown, and developmental contributions of the Pi-Mi complex beyond Mei3 induction have not been studied We are interested in how gene expression and molecular signaling come together to irreversibly establish the zygotic cell fate defined by mating repression and meiotic proficiency. Our current work employs genomics, genetics and biochemistry to directly addresses the above questions. As a basal eukaryote with powerful experimental tools, fission yeast is an excellent model system to understand mechanisms driving the zygotic cell fate switch, which is indispensable for sexual reproduction across phyla.
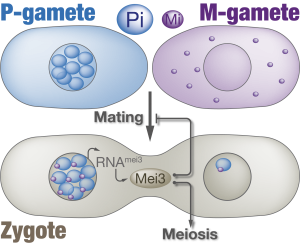
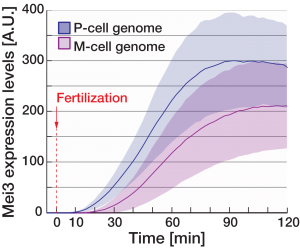 Figure 2: Mei3 rapidly induced from the P-parental genome
Figure 2: Mei3 rapidly induced from the P-parental genome