Aim of our research
- Cells live in changing environments. They have evolved complex signal transduction cascades to respond to these changes and mount an appropriate cellular response. Mitogen-Activated Protein Kinases (MAPK) cascades are conserved signal transduction modules shared among all eukaryotes. Budding yeast, Saccharomyces cerevisiae, has been a long-standing model system to study the regulation of these pathways. Single cell and quantitative measurements are needed to fully understand the dynamics of activation of these pathways. In the lab, our research focuses on three major aims:
– biosensor development
– Integration of multiple signals by MAPK pathways
– Regulation of gene expression -
Biosensors
In order to measure the signaling activity in live cells by microscopy, fluorescent reporters are required. Sometimes, endogenous proteins, which change cellular compartment when activated, can provide a good read-out of signaling activity. In situations where these reporters are not available, we have engineered synthetic proteins that display a relocation phenotype. We have done this to quantify MAP activity with a sensor name SKARS for Synthetic Kinase Activity Relocation Sensor. We have also engineered a dynamic Protein Synthesis Translocation Reporter (dPSTR) to monitor the kinetics of gene induction from specific promoters.
 Signaling activity biosensors. The SKARS sensor reports on the activity of the MAPK. Upon phosphorylation by the active kinase, the fluorescence relocates into the cytoplasm. The dPSTR reports on gene expression. Upon activation of the inducible promoter, the expression of a small peptide promotes the relocation of the fluorescent protein into the nucleus.
Signaling activity biosensors. The SKARS sensor reports on the activity of the MAPK. Upon phosphorylation by the active kinase, the fluorescence relocates into the cytoplasm. The dPSTR reports on gene expression. Upon activation of the inducible promoter, the expression of a small peptide promotes the relocation of the fluorescent protein into the nucleus.Integration of multiple signals by MAPK pathways
In nature, cells are often confronted with multiple signals that stimulate incompatible pathways. Therefore, cells, need to be able to take decisions in order to resolve this conflict. During development, nutrients can promote the growth and division of cells, while presence of a hormone will induce a cell-cycle arrest and differentiation of the cells. MAPK pathways are often implicated in these cell fate decision systems. We use budding yeast as a simple model eukaryote to understand the molecular mechanisms that are implicated in these cell fate choices.
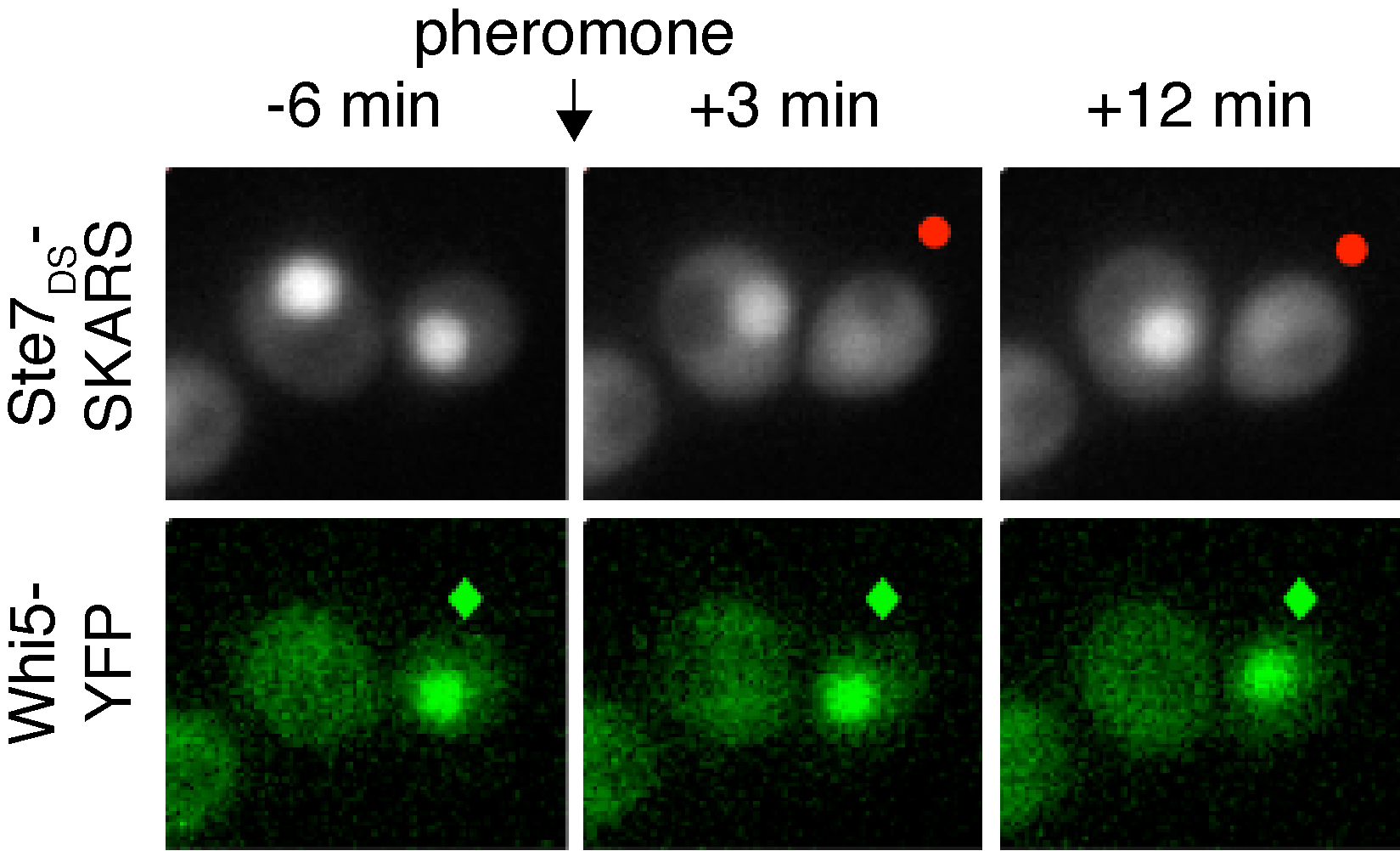 When yeast cells are stimulated with mating pheromone, they can either continue dividing or arrest their cell cycle to induce the mating MAPK cascade. This cell fate decision can be visualized in single cells by combining a SKARS reporter to monitor when cells activate the MAPK pathway and a cell-cycle marker (Whi5) to follow cell-cycle progression. In the images above note that only the cell in G1 (?) activates the MAPK pathway (•).
When yeast cells are stimulated with mating pheromone, they can either continue dividing or arrest their cell cycle to induce the mating MAPK cascade. This cell fate decision can be visualized in single cells by combining a SKARS reporter to monitor when cells activate the MAPK pathway and a cell-cycle marker (Whi5) to follow cell-cycle progression. In the images above note that only the cell in G1 (?) activates the MAPK pathway (•).Regulation of gene expression
Active MAPK can usually induce complex transcription programs implicating hundreds of genes. However, it remains poorly understood how a specific DNA sequence controls gene expression. More specifically, we don’t know how the number, the strength, the orientation and the position of transcription factor binding sites in the promoter sequence influence the level, the dynamics and the noise of mRNA production. Using single cell measurements, we can follow in real time the induction of gene expression and understand how the promoter architecture influence the expression output.
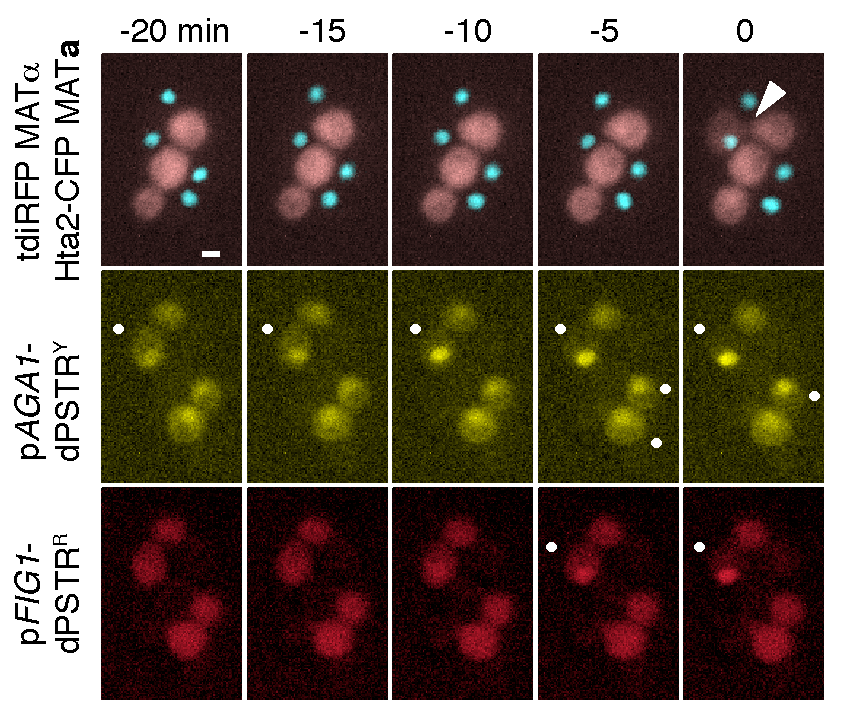 During the mating process between two haploid yeast cells, we can monitor when specific promoters are activated (•). By combining two dPSTR systems controlled by two different promoters pAGA1 and pFIG1, we observe that pAGA1 is induced in many cells. In contrast, pFIG1 is induced only in fusing cells shortly before fusion (arrow head).
During the mating process between two haploid yeast cells, we can monitor when specific promoters are activated (•). By combining two dPSTR systems controlled by two different promoters pAGA1 and pFIG1, we observe that pAGA1 is induced in many cells. In contrast, pFIG1 is induced only in fusing cells shortly before fusion (arrow head).
