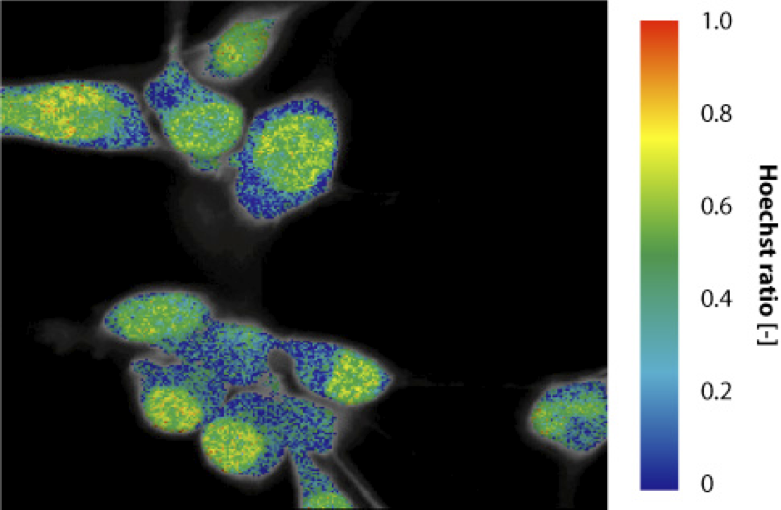
This algorithm allows to analyze fluorescence lifetime images. Global analysis allows to link some fitting parameter between all the pixels, while other will vary across the image. In the present case, the goal is to retrieve a well defined number of time constants and their relative contribution in each pixel. This optimization was tested on cells stained with calcein (live cell marker) and Hoechst (nucleic stain), which were image on our fluorescence lifetime imaging set-up. The FLIM image was fed to the optimization routine. The two time constants retrieved are 3.67 and 2.37 ns, compared to expected values of 3.85 and 2.35 ns. The ratio of Hoechst in the image gives a value close to zero in the cytoplasm and close to 50% in the nuclei where both dyes emit.
 Due to the large number of parameters to optimize simultaneously and the need to deconvolve the measured fluorescence decay, this fitting procedure can become extremely time consuming. We have devised a way to improve dramatically the time needed to reach convergence by generating an initial guess based on image segmentation.
Due to the large number of parameters to optimize simultaneously and the need to deconvolve the measured fluorescence decay, this fitting procedure can become extremely time consuming. We have devised a way to improve dramatically the time needed to reach convergence by generating an initial guess based on image segmentation.
This work has been published in the Biophysical Journal, S. Pelet, M. J. R Previte, L. H. Laiho andP. T. C. So, A fast global fitting algorithm for fluorescent lifetime imaging microscopy based on image segmentation, 2004, 87. 2807-2817.
The code for this algorithm is provided here as a zip archive: global_fit. The program has been written in Matlab 6.5. and requires the use of the optimization and image processing toolboxes.
