How can we reconstruct the evolutionary history of species with unprecedented accuracy? Nicolas Salamin and his team are pushing the boundaries of evolutionary biology through artificial intelligence. Their model, phyloRNN, uses deep learning to estimate molecular evolution parameters directly from DNA sequences — a major breakthrough that opens new avenues for biodiversity research.
Imagine attempting to reconstruct life’s evolutionary history by analyzing DNA sequences found in the genome of every individual. Traditionally, scientists have relied on mathematical models, yet these often rest on simplifying assumptions that may fall short of capturing the full complexity of evolution. To address this challenge, Nicolas Salamin and colleagues Daniele Silvestro (ETH Zurich) and Thibault Latrille (UNIL) have developed a novel approach that harnesses the power of artificial intelligence — particularly deep learning.
They have developed a new artificial intelligence model, called phyloRNN (github.com/phylornn/phylornn), designed to directly analyze multiple DNA sequence alignments and estimate key parameters of molecular evolution — such as the rate at which different genomic regions change and the overall divergence that has occurred — all without relying on a pre-existing evolutionary tree. The development of phyloRNN introduces a novel strategy, combining numerical simulations of genome evolution with a supervised deep learning model (see figure).
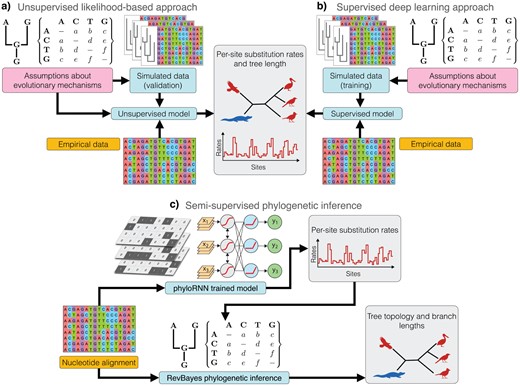
Essentially, the researchers generated large volumes of synthetic data that mimicked real-world scenarios — including complex patterns of rate variation that are notoriously difficult to model using traditional mathematical approaches. These simulated datasets were then used to train the phyloRNN model, enabling it to learn intricate relationships between DNA sequence patterns and the underlying evolutionary processes.
The predictions made by the phyloRNN model regarding evolutionary rates proved to be as accurate — and in many cases significantly more accurate — than those obtained through traditional methods, especially in complex evolutionary scenarios. The researchers didn’t stop there: they demonstrated how these AI-powered estimates could be integrated back into conventional phylogenetic frameworks to enhance the accuracy of tree reconstruction. By incorporating site-specific evolutionary rates predicted by phyloRNN into a Bayesian framework, they observed a substantial improvement in phylogenetic inference, notably in the estimation of branch lengths.
This innovative semi-supervised approach, combining the strengths of deep learning for rate estimation with the rigor of probabilistic inference for tree construction, points to a promising future for phylogenetic analysis. It enables the integration of more flexible and realistic models of evolution. This research paves the way for further advances and collaborative efforts across computational biology, computer science, and evolutionary studies. The potential of deep learning in phylogenetics continues to inspire new innovations and explorations within this interdisciplinary field, contributing to a deeper understanding of the dynamics and mechanisms driving species evolution and biodiversity.
Professor Nicolas Salamin is a biologist who incorporates modelling and artificial intelligence into his research to understand the mechanisms leading to the evolution of species and biodiversity.
Faculté de biologie et de médecine
Modeling, Deep Learning, Evolutionary genomics
