FAIRShare Explorer @DSBU, developed by the DSBU, helps researchers at FBM UNIL and CHUV identify the most appropriate, standards-compliant data repository for their datasets.
This tool is built on a comprehensive analysis of repositories recommended by major scientific publishers (e.g., Nature Publishing Group, PLOS, etc.) and has been specifically adapted to the data types commonly produced or analyzed within the FBM UNIL–CHUV environment.
The selection focuses on repositories relevant to data generated in Life Sciences, Biomedical, and Clinical Research, aligning with the disciplines of the Faculty of Biology and Medicine, including both the Fundamental Science Section and the Clinical Science Section.

Access FairShare Explorer Web App
FairShare Explorer is directly accessible to all users who have already access to DataSquid—you’ll find it as a dedicated board within your catalog space.
For those who do not yet use DataSquid or do not have an active setup, FairShare Explorer is also available externally without a login, as long as you are connected to the UNIL or CHUV network environment.
⚠️ Important: If you are working from outside the university or hospital, make sure to connect via VPN to access the tool.
Link to FAIRShare Explorer ©DSBU: https://dsbu.unil.ch/datasquid-fbm/repositories
Best Practices for Using FairShare Explorer
- Integrate Data Sharing into Your DMP Early Embed your data sharing strategy within your Data Management Plan (DMP) from the outset to ensure a clear roadmap for describing, preserving, and publishing your data.
- Prioritize European, Public, and Domain-Specific Repositories To safeguard long-term access and guard against repository closures or policy shifts, choose trusted, non-commercial repositories hosted in Europe. Wherever possible, select repositories that are specific to your discipline or data type—these often provide superior visibility, tailored tools, and optimized support for large or complex datasets.
- Ensure FAIR Compliance and Contingency Planning Only use repositories that implement the FAIR principles (Findable, Accessible, Interoperable, Reusable) and maintain robust contingency measures—such as mirrored archives—to guarantee data preservation over time.
- Use Filters for Precision Leverage FAIRShare Explorer’s filtering options (discipline, data categories and subcategories, confidentiality level) to pinpoint the repository that best matches your dataset’s characteristics and access requirements.
- Explore Specialized Subcategories Dive into the Explorer’s subcategory listings to find the most precise repository fit—whether it’s a niche archive for imaging, omics, neuroscience, or clinical data.
- Adhere to Legal and Ethical Standards For human-subject or sensitive datasets, confirm that your chosen repository meets all applicable legal, ethical, and institutional requirements before depositing data.
- Document Thoroughly and License Openly Provide rich metadata and detailed README documentation to maximize discoverability and reuse. Apply an open license (e.g., CC BY 4.0) whenever possible to enable broad data sharing while ensuring proper attribution.
Reminder: all data supporting a publication (raw, processed, and analytical) must also be archived on long-term storage provided by DCSR-UNIL (LTS) or DSI-CHUV.

What you’ll find in the Explorer
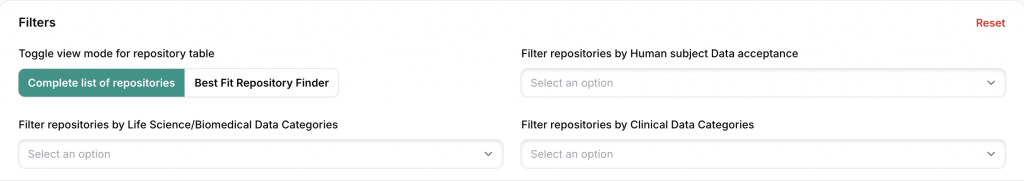
Two Main Navigation Modes
Complete repository view
- An exhaustive list of all repositories included in the system.
- list of repositories with key informative details for efficient submission: data types and formats, submission process, metadata standards, sharing licenses, and more

Comparative view: “Best Fit Repository Finder”
Allows for comparing repositories based on defined criteria (data type, discipline, legal requirements…) to highlight the most suitable ones.
- Quickly identify:
- Recommended repositories (green)
- Repositories to avoid (red), based on the criteria above
- Official recommendations from the FBM Dean’s Office and DSBU
- Priority is given to:
- European repositories, when an equivalent exists outside Europe.
- Specialized repositories, which are better suited than generalist ones for complex or sensitive datasets.
- Priority is given to:
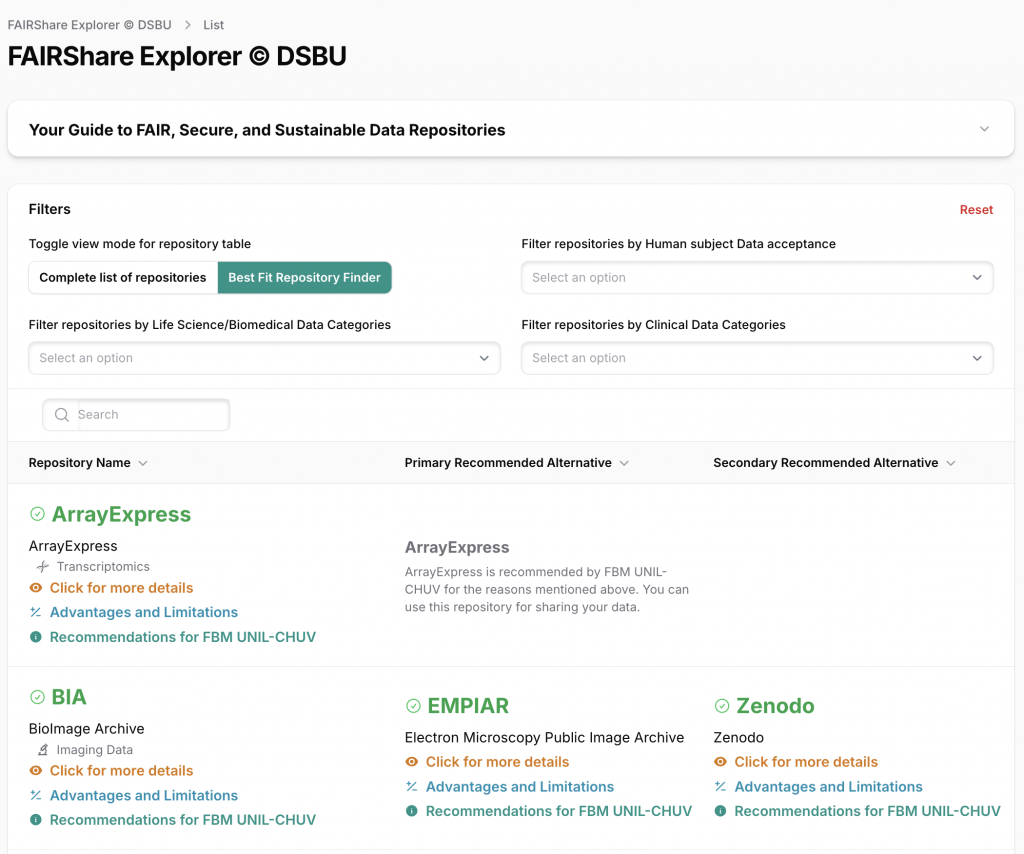

Filters Tailored for UNIL–CHUV Researchers
FairShare Explorer includes advanced filters to:
- Sort repositories by discipline, data categories (e.g., imaging, omics, clinical) and subcategories
- Identify platforms that accept human subject data, whether:
- De-identified
- Non-anonymized under specific conditions
- Sort repositories by name or keyword
These filters guide you toward ethically sound, legally compliant, and publication-ready choices
Filters repositories by name or keyword

Filters by Discipline and Data Type
Two main disciplinary categories are used to refine the search:
🔬 1. Life Science / Biomedical Data Categories

Includes data from Life Science and Biomedical research, such as:
- Genomics (e.g., DNA sequencing)
- Transcriptomics
- Proteomics
- Epigenomics
- Electron microscopy
- Imaging
- In vivo Imaging (from animal models)
- Flow cytometry
- Spectroscopy
- Behavioral data
- Electrophysiology data
🏥 2. Clinical Data Categories
Covers data from studies involving human participants.

Clinical data classification is aligned with European standards such as CDISC SDTM:
- Demographic :Data Background information on participants:
- Clinical Data : Medical and health-related information:
- Interventional : Data Details of treatments and procedures:
- Outcome Data : Measures of intervention results
- Patient-Reported Data: Health information provided directly by patients:
- Administrative Data: Data for compliance and study tracking:
- Claims Data: Healthcare utilization data:
- Electronic Health Records (EHR) Data: Digitally collected clinical information:
- Medical Imaging Data
Note: Some human-origin data (e.g., genomics, imaging) may fall under Life Science / Biomedical but still require secure sharing practices.
Navigating by Data Subcategories
To fine-tune your results, FairShare Explorer provides an additional “Data Subcategories” filter.
Guided Selection by Data Subcategory

- Select a main Data Category (e.g., Genomics)
- View all related subcategories—such as DNA sequencing, DNA polymorphisms, and more.
- Pick one or several subcategories, or click “Select All” to include every subcategory within that domain.
Example:
To find a repository for DNA sequencing data:
- Select the “Genomics” category
- Choose the “DNA sequencing” subcategory
- You’ll then see:
- In the Complete List: all compatible repositories
- In the Comparative View: recommended repositories e.g., ENA (Europe) is favored over SRA (US) due to European data sovereignty.
Complete List

Comparative View

Filters for Human Data Sharing ( Anonymized or Not)
To securely share human subject data, FairShare includes two essential filters:
- Accepts De-Identified Human Sample Data
- Accepts De-Identified Human Subject Data
- Accepts Non-Anonymized Data

Accept De-identified Human Sample Data and Subject Data
- For anonymized or de-identified data
- Requires explicit informed or general consent for open data sharing
- Compatible with many repositories, including generalist ones like Zenodo
Accept Non Anonymized Data

- For non-anonymized personal or sensitive data
- Must be shared under restricted access
- Recommended repositories:
- EGA (European Genome-Phenome Archive)
- Horus Dataset Catalogue CHUV A secure restricted-access repository at CHUV where only metadata is public (e.g., on Zenodo). Access is granted via a Data Transfer and Use Agreement (DTUA), following the SPHN template.
What You’ll Find for Each Repository
Since information available on repository websites is often fragmented or unclear, FairShare Explorer restructures and enriches it, offering a clear, practical, and FAIR-aligned overview.
Advantages and Limitations Section
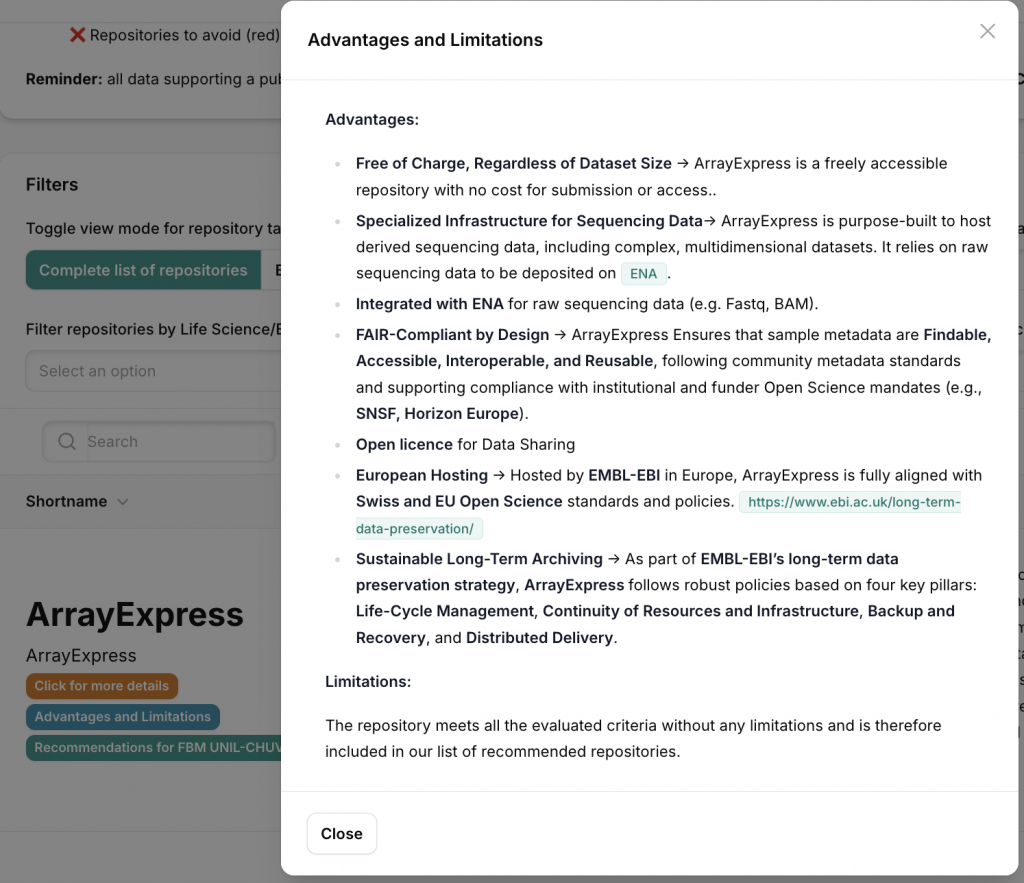
This section provides a balanced assessment of the repository, highlighting both its strengths and limitations. The advantages typically include technical features (e.g., storage capacity, metadata support, integration with other platforms), policy alignment (e.g., FAIR principles, licensing options), and practical benefits for users (e.g., cost-free access, long-term archiving). The limitations address areas where the repository may fall short, such as geopolitical issues, lack of domain-specific tools, restricted support for complex file formats, or absence of certain preservation guarantees. Together, these elements help assess whether the repository is suitable for specific research needs and compliance requirements.
Detailed profile

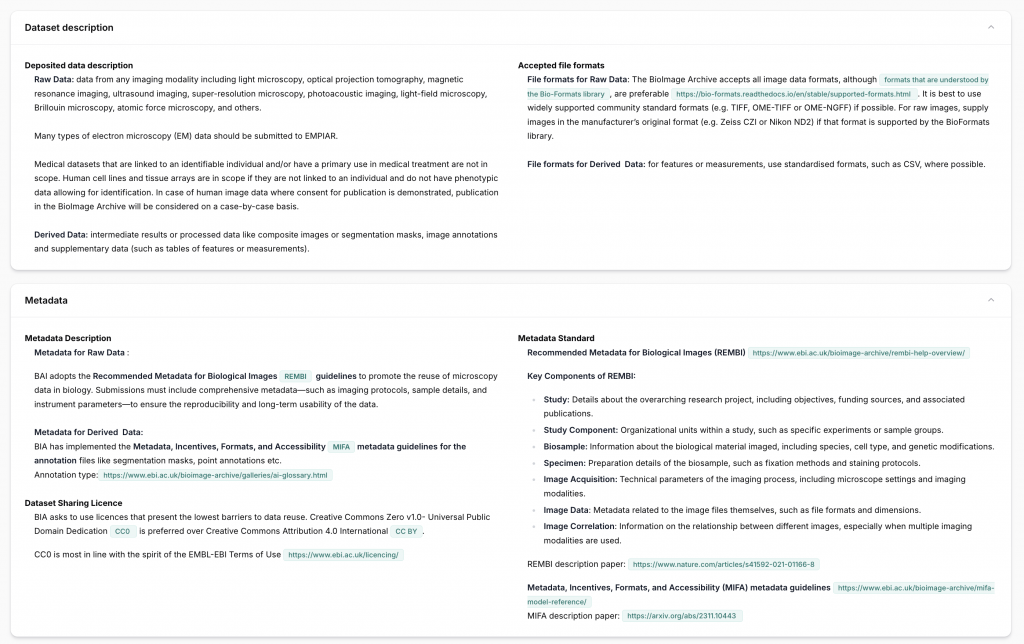
Every repository entry includes a detailed profile, covering:
- Repository Name and Short Name The full name and an acronym or concise identifier for quick reference.
- Type (Primary Repository, Database, Catalog, Aggregator) Clarifies whether the repository stores datasets directly (Primary Repository, Database), indexes them (Catalog), or aggregates from other sources.
- Linked Data Categories and Subcategories Shows the types of data accepted (e.g., imaging, genomics, proteomics) and relevant subdomains (see our related official list).
- Acceptance of Sensitive/Human Data Indicates whether the repository accepts de-identified or, in specific cases, non-anonymized human data, along with ethical requirements.
- Main URL and Description Includes a direct link and a short description of the repository’s mission and scope.
- Size Limits for Deposits States any constraints on individual file sizes or total dataset volume.
- Types of Data Accepted and File Formats Lists supported data types and formats (e.g., FASTQ, mzML, DICOM), ensuring compatibility.
- Metadata Description and Supported Standards Specifies what metadata is required and which community standards are supported (e.g., ISA-Tab, MIAME, MIAPE).
- Server Location and EU/non-EU Status Identifies where the data is physically hosted, which is relevant for GDPR compliance.
- Equivalent European Repositories (if applicable) Suggests trusted EU-based alternatives to non-European repositories to mitigate legal or ethical risks.
- Licensing Options and Usage Rights Lists available licenses (e.g., CC BY 4.0, CC0) and any associated reuse conditions requested by the repository.
- Profit/Non-Profit Status Clarifies whether the repository is operated by a non-profit organization or a commercial provider.
- FAIR/SNSF Compliance Indicates whether the repository supports FAIR principles and meets Swiss National Science Foundation (SNSF) and Horizon Europe requirements.
- re3data and FAIRsharing Links Provides external validation and metadata via links to recognized registries.
- Replication Policies Explains whether the repository mirrors or replicates data to ensure redundancy and preservation.
- Cross-Referencing with Other Databases Indicates integration with external repositories or databases (e.g., UniProt, GEO), which enhances data visibility and interoperability.
- Business and Contingency Plans Provides insight into long-term sustainability, including backup plans, governance, and funding continuity.

Repository Usage Recommendations
Each repository is accompanied by a tailored set of recommendations to help you:

Guidance for repository usage
- Understand the Key Benefits → Repositories are evaluated for:
- Cost-free deposition (even for large datasets)
- Specialized infrastructure (e.g., for imaging, genomics)
- Built-in FAIR compliance
- EU hosting
- Integration with other scientific resources
- Follow File Format & Metadata Standards → Recommendations include accepted file types and schemas from the repository. → Use DataSquid to automatically generate:
- Structured README files
- Metadata conforming to REMBI, ISA-Tab, MIAPE, etc.
- Choose the Right Repository → Prioritize data-type-specific repositories for better visibility, discoverability, and compliance.
- How to publish your dataset
Data Sharing Licence
- Apply the Right Licence → FBM recommends Creative Commons Attribution (CC BY 4.0) for open and FAIR-compliant sharing. → Aligned with UNIL Directive 4.5 and Directive institutionnelle « Recherche sur l’être humain » du CHUV.
- When the repository mandates the Creative Commons Zero v1.0 Public Domain Dedication CC0, consequently, all researchers at FBM/CHUV must deposit their data under CC0.
- Sensitive and non-anonymized datasets from CHUV must be shared under a restricted access model, governed by a Data Transfer and Use Agreement (DTUA). In such cases, if the Principal Investigator and the relevant committee approve the request, a collaboration agreement may be established. The dataset can then be transferred to the external party under the terms of a CHUV DTUA, which is based on the standard template provided by the Swiss Personalized Health Network (link).
Data to be archived in Long Term Storage (LTS)
- Preserve Your Data in Long-Term Storage → Archive raw, processed, and published datasets at:
- DCSR–UNIL (LTS) or
- DSI–CHUV, in accordance with institutional requirements.
European Equivalent Repository
- Use the “Best Fit Repository Finder” → This feature suggests EU-based alternatives when the original repository is outside Europe, helping you stay compliant with Open Science and data protection regulations.
Institutional Support
- DSBU support
- FBM Core Research Facilities
Get Support from the DSBU
The DSBU team offers personalized support throughout the data deposition process https://wp.unil.ch/dsbu/sharing/:
- Tutorials and short, hands-on training sessions
- Guidance on preparing your dataset for sharing
- Tools and templates to help produce a structured README file and Enriched Metadata describing data types, acquisition methods, instrumentation, software, etc. using DataSquid → https://wp.unil.ch/dsbu/tools_readme/
- Support in ensuring metadata quality and compliance with FAIR principles
- Step-by-step assistance with submission
- Advice on data reuse, copyright, and licensing
- Support with secondary data deposition on institutional infrastructure, ensuring secure long-term preservation through tape-based archiving.