Experiments in microbiology are typically conducted in well-mixed liquid cultures, as it is important to abstract out some of the complexity of the real world. This is no different for our lab. Yet, we know that microbes mostly live in spatially-structured biofilm, so part of the lab tries to understand how spatial structure can affect ecological and evolutionary dynamics in microbial communities.
One aspect of this work is to understand how a community will assemble over the length of a tube, such as microfluidic device. We have developed mathematical models that map out interactions across space as a function of the changing substrate concentrations as one species grows and changes its local environment [1]. We are now testing some of these ideas using experiments.
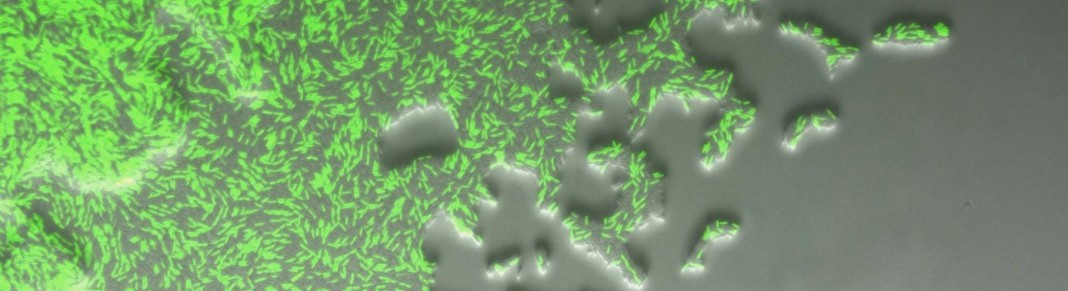
A confounding factor when studying spatial structuring is understanding to what extent metabolic interactions (e.g. how consuming or producing metabolites affects others) versus physical interactions (e.g. to what extent a species sticks to another one) affect spatial patterning. We are studying this question using different species combinations in colonies growing on agar plates.
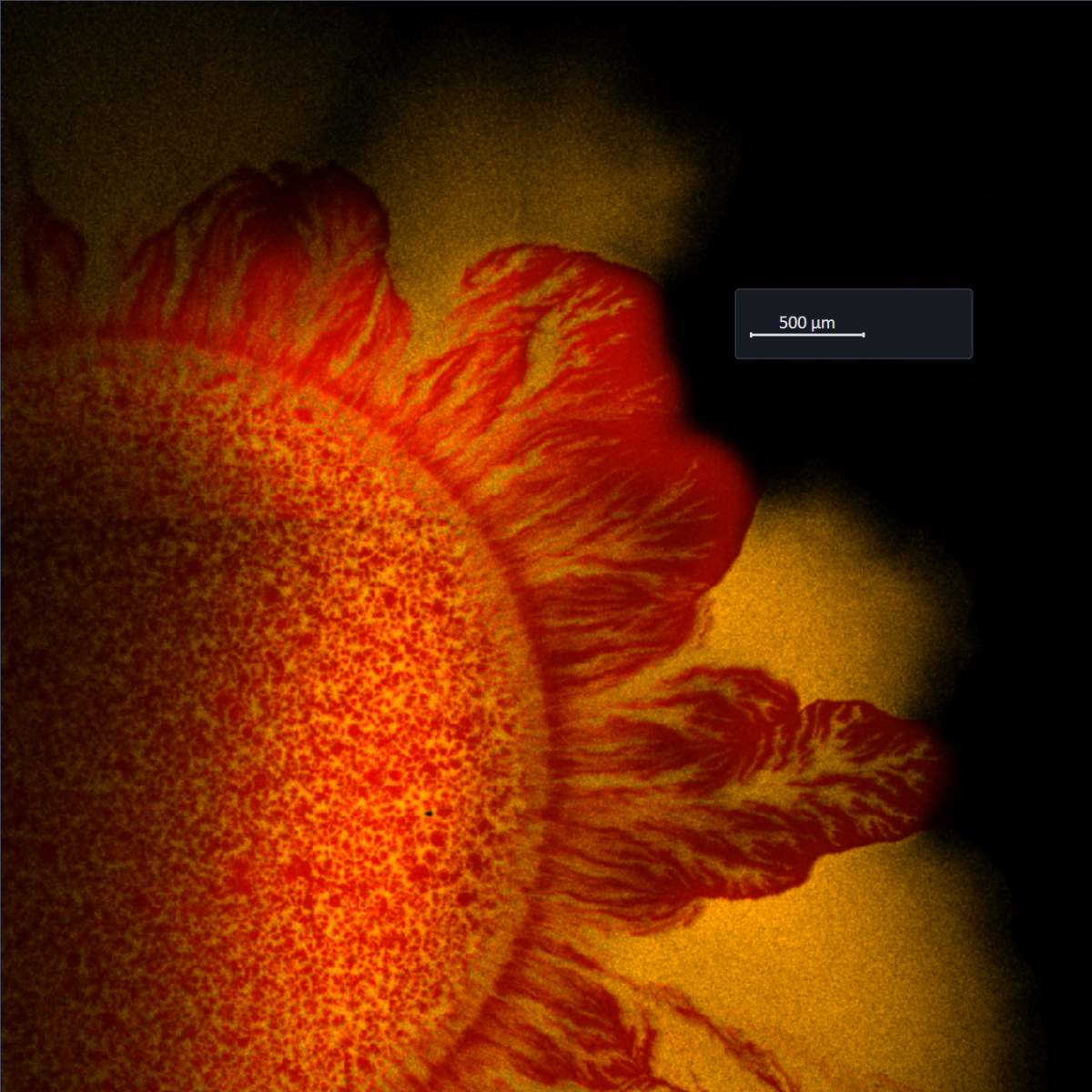
In the past, we have also studied how phage infections can be affected by spatial patterning in colonies composed of a strain that is sensitive to the phage and another that is resistant to it. We found that pockets of low phage infectivity could arise, but mainly due to slow growth [2].
References
1. O. Meacock, S. Mitri (2023) bioRxiv.
2. S. Testa, S. Berger, P. Piccardi, F. Oechslin, G. Resch, S. Mitri (2019) Comm Biol.