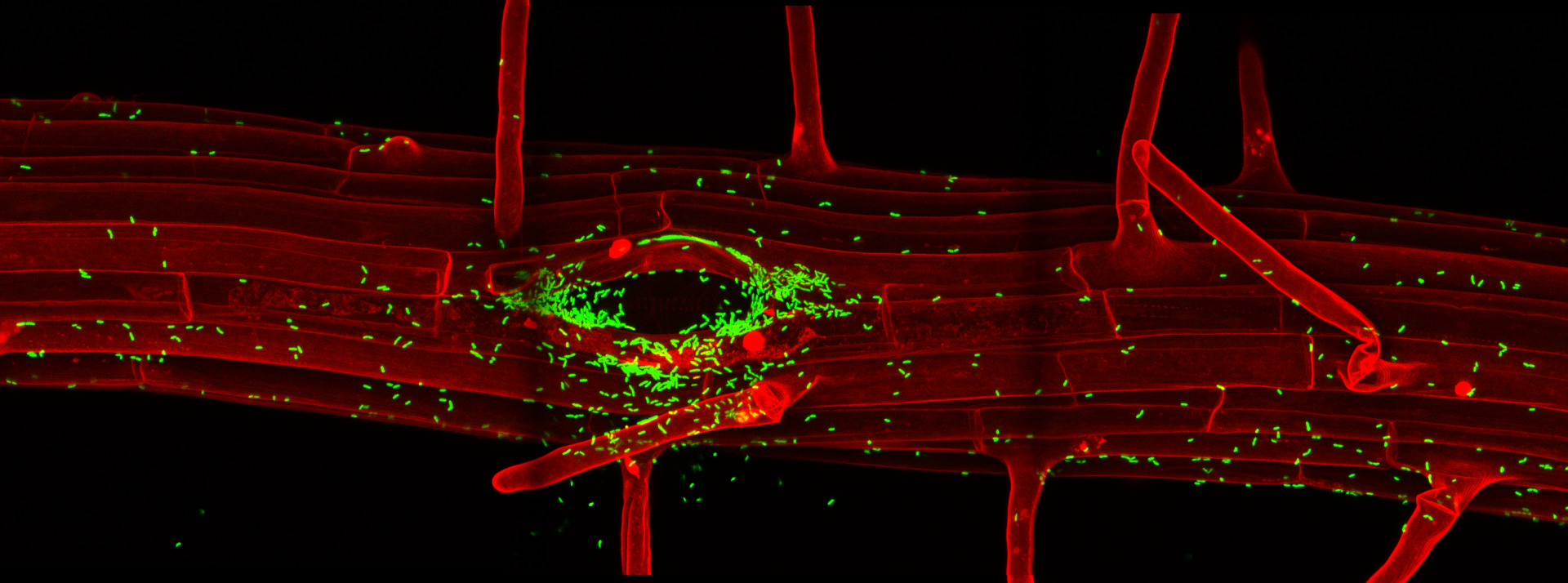
Wave lines: Mapping the complexity of the plant membrane system
 Eukaryotic cells maintain a dynamic and complex network of membrane compartments, such as the endoplasmic reticulum, Golgi apparatus, endosomes, and vacuole/lysosomes. These compartments separate and organize proteins within the cell and impose temporal as well as spatial restrictions on protein activities. Compartment identities are maintained in spite of a constant exchange of material. This process of continuous generation and consumption of compartments defies any simple analogies with static macroscopic structures and membrane compartments should rather be compared to waves, whose very essence is the dynamic flow of material through them.
Eukaryotic cells maintain a dynamic and complex network of membrane compartments, such as the endoplasmic reticulum, Golgi apparatus, endosomes, and vacuole/lysosomes. These compartments separate and organize proteins within the cell and impose temporal as well as spatial restrictions on protein activities. Compartment identities are maintained in spite of a constant exchange of material. This process of continuous generation and consumption of compartments defies any simple analogies with static macroscopic structures and membrane compartments should rather be compared to waves, whose very essence is the dynamic flow of material through them.
The complexity of the membrane compartments of plant cells are only insufficiently appreciated, especially for cells in the context of the whole plant. One of the problems is that many membrane compartments have an inconspicuous morphology, especially at the current resolution of fluoresent live-imaging. Therefore, they can only be distinguished by co-localisation with spectrally distinct fluorescent markers. At the Salk Institute, we developed a vector series for plant transformation for a non-Gateway-based recombination . This vector set allows the use of a recently developed Arabidopsis ORF library for large scale generation of fluorescent fusion constructs in plants. These vectors are an alternative to Gateway. Their CRE/lox system is non-patented, cheap and compatible with the Arabidopsis ORF collection, currently encompassing about 13.000 clones. We used this system (Liu et al., 1998) to generate a large set of membrane compartment markers. This set allows the definition of compartments and trafficking pathways in a developmental context and does not rely on transient expression in protoplasts or cultured cells. Our set of transgenic plants with yellow, red and blue variants of compartment markers is now established and has been published (Geldner et al., 2009).
Wave constructs and seeds are available at the Nottingham Arabidopsis Stockcenter (NASC)
American researchers can order from there while the stock are being transferred to the ABRC.
When using our Wave construct and lines or our pNIGEL vector series, please cite our paper in The Plant Journal:
Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J.Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set.Plant J. 2009 Jul;59(1):169-78. Epub 2009 Feb 26. PMID: 19309456
| Wave number | gene name | AGI | EYFP (Yellow) | mCherry (Red) | mCerulean (blue) | mTFP1 (bluegreen) |
|---|---|---|---|---|---|---|
| 1 | tag only (free FP) | n.a. | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 2 | RabF2b (ARA7) | At4g19640 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 3 | Rab C1 | At1g43890 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 5 | RabG3f | At3g18820 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 6 | NIP1;1 | At4g19030 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 7 | RabF2a (Rha1) | AT5g45130 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 9 | VAMP711 | AT4g32150 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 11 | Rab G3c | At3g16100 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 13 | VTI12 | At1g26670 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 18 | Got1p homolog | At3g03180 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 22 | SYP32 | At3g24350 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 24 | Rab A5d | At2g31680 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 25 | Rab D1 | At3g11730 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 27 | Rab E1d | At5g03520 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 29 | Rab D2a | At1g02130 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 33 | Rab D2b | At5g47200 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 34 | Rab A1e | At4g18430 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 127 | MEMB12 | At5g50440 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 129 | Rab A1g | At3g15060 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 131 | NPSN12 | At1g48240 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
| 138 | PIP1;4 | At4g00430 | DNA/seeds | DNA/seeds | DNA/seeds | DNA only |
FOR THE LATEST, UPDATED INFORMATION ON THE WAVE LINES, DOWNLOAD THIS PDF:
![]() Wave_line_info_sheet (826 Kb)
Wave_line_info_sheet (826 Kb)
The pNIGEL recombination vector series
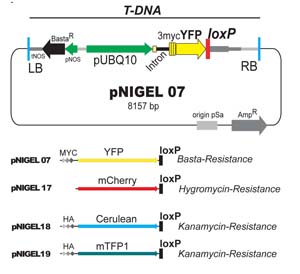 The pNIGEL vectors were developed to allow optimal co-localisation between lines carrying different fluorescent protein fusion constructs. The three fluorescent proteins chosen can be easily separated spectrally. The Cerulean and mTFP1 vectors contain an additional 3xHA tag, the EYFP vector contains an additional 3xMyc epitope. Vectors of different colours carry different resistances, such as to allow selection of crossing products and/or sequential transformations with different constructs. Marker expression is driven by the intron-bearing, endogenous UBQ10 promoter, in order to promote stable expression levels and minimise overexpression artifacts while obtaining sufficiently strong signals. pNIGEL vectors are based on the pGREEN II vector series, in which the bacterial Kan Resistance was exchanged against Amp Resistance. For additional information, please refer to the publication (Geldner et al., 2009).
The pNIGEL vectors were developed to allow optimal co-localisation between lines carrying different fluorescent protein fusion constructs. The three fluorescent proteins chosen can be easily separated spectrally. The Cerulean and mTFP1 vectors contain an additional 3xHA tag, the EYFP vector contains an additional 3xMyc epitope. Vectors of different colours carry different resistances, such as to allow selection of crossing products and/or sequential transformations with different constructs. Marker expression is driven by the intron-bearing, endogenous UBQ10 promoter, in order to promote stable expression levels and minimise overexpression artifacts while obtaining sufficiently strong signals. pNIGEL vectors are based on the pGREEN II vector series, in which the bacterial Kan Resistance was exchanged against Amp Resistance. For additional information, please refer to the publication (Geldner et al., 2009).
The pNIGEL vector series can also be ordered at NASC.
Sequence information can be downloaded from Genbank.
For a complete list of pNIGEL vectors, please refer to this pdf file:
![]() pNIGEL_vector_series (1200 Kb)
pNIGEL_vector_series (1200 Kb)
Wave constructs are produced by recombination of a given pNIGEL with the respective ORF, cloned into the pUNI51 vector by the SSP consortium. Below is shown part of the resulting fusion construct using the Wave131Y construct as example (NPSN12 ORF fused to pNIGEL07):
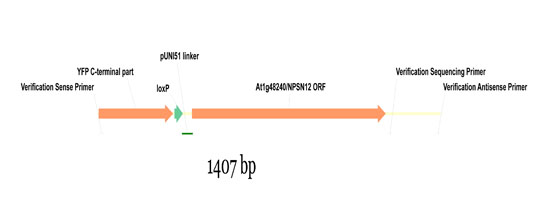
Example sequences of recombined Wave constructs can be downloaded below as genbank files. In order to obtain the exact sequence of your construct of interest, just exchange the example NPSN12 ORF with your ORF of interest (including start and stop codon).
![]() Wave131_in_pNIGEL_07__YFP_.gb (94 Kb)
Wave131_in_pNIGEL_07__YFP_.gb (94 Kb)
![]() Wave131_in_pNIGEL_17__mCherry_.gb (87 Kb)
Wave131_in_pNIGEL_17__mCherry_.gb (87 Kb)