Genome maintenance and DNA defense
Genome maintenance and DNA defense are essential for life. Our genetic material is constantly challenged—by errors during DNA replication and chromosome segregation, by environmental damage, and by invasive elements like viruses and plasmids. Without systems to repair, maintain, and defend DNA, cells would quickly lose the integrity of their genomes, leading to disease, malfunction, or death. By uncovering the molecular mechanisms behind these protective processes, we can better understand how life safeguards its most valuable blueprint: the genome.
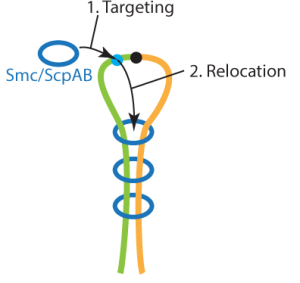
Key factors in chromosome folding: The SMC ATPase complexes
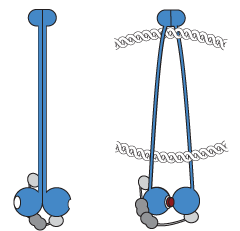
Our research focuses on the multi-subunit SMC ATPase complexes, which play a crucial role in organizing chromosomes by forming DNA loops, thus bringing together distal segments of chromosomes. These complexes are ATP-powered DNA motors that actively extrude DNA loops; they take large steps along the DNA and are able to bypass obstacles, while also maintaining directionality over long periods of time. To gain a better understanding of the mechanisms behind this process, we are studying bacterial SMC complexes and the yeast Smc5/6 complex using biochemical techniques and cryo-electron microscopy at the Dubochet Center for Imaging in Lausanne (@DCI_EM). Our work has led to the development of the DNA segment capture model, which we are now testing.
SMC Wadjet systems in bacterial immunity
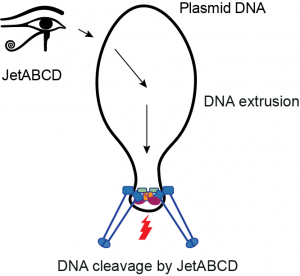
Wadjet systems are derivative SMC complexes that play a role in bacterial immunity, rather than in chromosome folding. We have recently shown that Wadjet/JetABCD eliminates plasmids by cleaving circular DNA molecules. We are interested in understanding how the Wadjet complexes specifically recognize and target smaller circular DNA molecules, while sparing the host chromosome from processing. The eukaryotic Smc5/6 complexes have been shown to play an apparently related role in defense against infection by viruses (HBV, EBV and others). We are working to understand how these activities in viral and plasmid defense are related to one another and to the more canonical functions of SMC complexes in genome folding and maintenance. Recently, we have also started investigating other bacterial defense systems that act on DNA and share similarities with chromosomal DNA repair machinery.
ParABS & cellular regulation
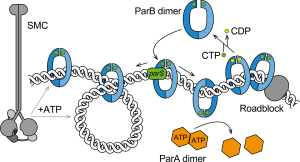
ParABS systems are important for promoting chromosome segregation and plasmid maintenance in many bacteria and some archaea, as well as for supporting additional regulatory functions within the cell. These systems work by binding ParB proteins to centromeric parS sequences to form large nucleoprotein complexes that interact with specific ATPases (ParA and Smc) to partition plasmid and chromosome copies. We have recently discovered that ParB proteins are enzymes – the first known CTP hydrolases – which form DNA sliding clamps that self-load onto parS DNA. We are particularly interested in how the combination of ParB CTP binding and hydrolysis with ParA and Smc ATPases promotes chromosome organization and segregation in bacteria.
Our approach
A deep understanding of the architecture and structure of macromolecular assemblies is often essential for gaining a mechanistic understanding of their function. Our research combines genetics, molecular and cell biology, biochemistry, and structural biology to reveal the molecular basis of protein function and cellular activity. We use a range of biophysical techniques and structural biology to investigate protein-DNA complexes and DNA organization in vitro, as well as in vivo techniques such as ChIP-Seq, Hi-C, directed or random mutagenesis, and site-specific cross-linking. By combining these approaches, we hope to gain insights into the underlying mechanisms of these complex systems.
New focus?
We are always open to exploring new areas of research, including the roles of other SMC and SMC-like proteins (such as Rad50 and RecN) in maintaining DNA integrity in both bacteria and eukaryotes.
