Bacterial Cell Biology
Our lab is mainly focusing on the biology of bacterial genomes using two model Alphaproteobacteria: the Caulobacter crescentus environmental bacterium with a unique circular chromosome and the Agrobacterium tumefaciens plant pathogen with a multi-partite genome.
- Chromosome replication and maintenance in Alphaproteobacteria
Over the last decade, we characterized a diversity of regulatory mechanisms ensuring that the bacterial genome is replicated at the right time of the cell cycle, at the right frequency and with the highest fidelity. We recently showed that the replisome of Caulobacter crescentus is spatially associated with its DNA mismatch repair (MMR) system, accelerating the detection and the repair of replication mistakes. In this bacterium, the regulated inactivation of the DnaA (RIDA) replication initiator appears to be the main mechanism controlling the frequency of chromosome replication and it is the target of a few novel regulatory pathways adapting cell cycle progression to environmental conditions. We are now exploring how Alphaproteobacteria with multi-partite genomes, such as Agrobacterium tumefaciens, control the replication of more complex genomes.
- Epigenetic control of the cell cycles of Alphaproteobacteria
Our past work uncovered a strong connection between DNA methylation by the conserved adenine methyltransferase CcrM and cell cycle control in Caulobacter crescentus. We are now investigating how adenine or cytosine methylation can influence gene expression and wish to identify novel and conserved epigenetic regulators of the cell cycle in Caulobacter crescentus and Agrobacterium tumefaciens.
- The impact of DNA methylation on genome maintenance/evolution in Alphaproteobacteria
When DNA methyl-transferases (MTases) are associated with cognate restriction enzymes in so-called restriction-modification systems (RMS), they are predicted to play a role in limiting horizontal gene transfers and thus in promoting genome manintenance. Still, the methylation of cytosines can also accidentally increase the occurence of C-to-T mutations at methylated DNA motifs and can thus become a source of evolution. We are currently exploring the impact of RMS and cytosine MTases on the evolution/maintenance of Alphaproteobacterial genomes.
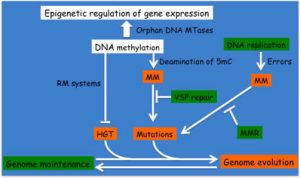
Schematic highlighting the impact of DNA methylation on genome evolution/maintenance and on gene expression.
(MTase: methyltransferase; RM: Restriction/Modification; MM: DNA mismatch; VSP: Very Small Patch; MMR: DNA Mismatch Repair; HGT: Horizontal Gene Transfer; 5mC: 5-methyl-cytosine)