Our aim is to decipher the molecular mechanism of bacterial growth
Understanding this process is a fundamental goal of microbiology and is directly linked to the synthesis and remodeling of the surface layers surrounding bacterial cells. These surface layers are of critical importance to bacterial survival by mediating protection against osmotic rupture, determining cell shape, while posing formidable permeability barriers. Thus, cell envelopes are a highly effective target for antibiotics, but conversely also significant contributors to antibiotic resilience and resistance.
Bacterial cell envelope architecture and complexity varies depending on the species, but generally includes a cytoplasmic inner membrane surrounded by a peptidoglycan cell wall layer (e.g. Gram-positive). Di-derm bacteria contain an additional outer membrane layer (Gram negative) and glycan polymers in Acid-fast bacteria. In our lab we use the classical di-derm model organisms Escherichia coli (Gram-negative) and Corynebacterium glutamicum (Acid-fast) to study these processes.


In our research we seek to understand the fundamental spatiotemporal mechanisms underlying the biogenesis of bacterial cell envelopes and characterize their biophysical properties using an interdisciplinary approach combining bacterial genetics, imaging (fluorescence microscopy, cryo-electron tomography and atomic force microscopy), biophysics, and chemistry.
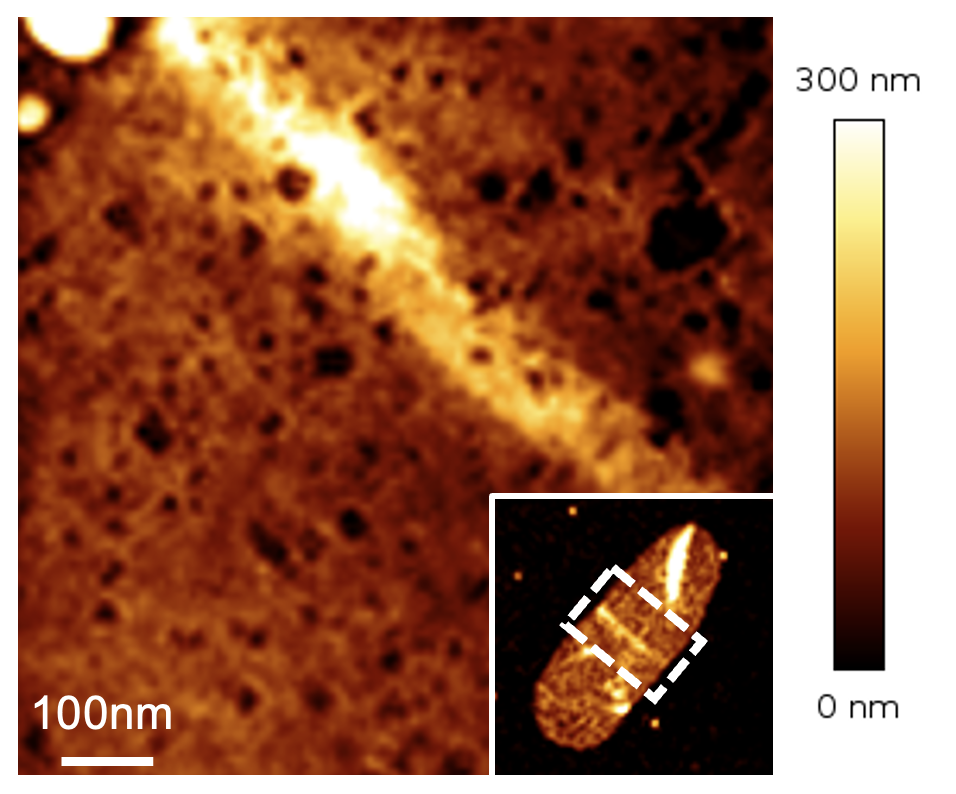
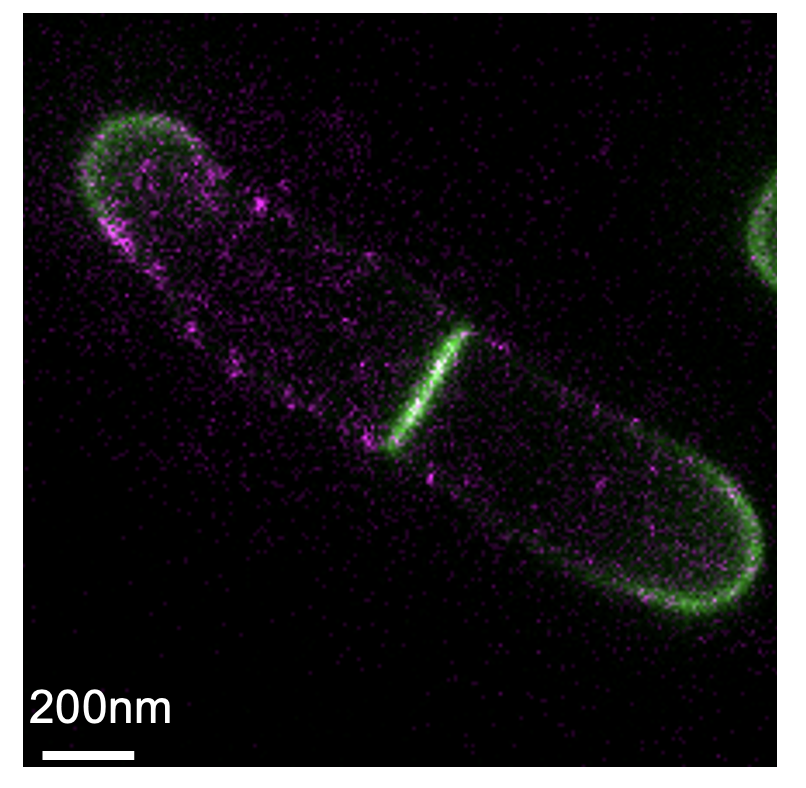
mycomembrane porin PorH (magenta) in C. glutamicum
