We have developed an advanced documentation solution, Datasquid, designed to enhance reproducibility in research by automating the process of data documentation for each experiment. The tool seamlessly integrates with local or institutional equipment database and internal laboratory protocol to generate detailed README files for individual experiments. By capturing all relevant metadata and procedural information, DataSquid ensures that each experiment is thoroughly documented, promoting consistency, transparency, and replicability across research workflows.
To ensure proper implementation and optimal use of DataSquid, PIs must first contact our team. We will assist in setting up and customizing the platform to meet the specific needs of your laboratory. Additionally, we offer personalized training sessions for lab members, ensuring that everyone is well-prepared to utilize DataSquid effectively and efficiently in their research workflows.
Link to DataSquid: https://dsbu.unil.ch/datasquid
Key Functionalities:
Automatic Documentation Generation:
- Data Management Plan (DMP)are automatically generated for documenting your NAS Projects.
- Individual README files are automatically generated for data folders stored on the NAS-DCSR.
- A global README file is produced for TARs documentation before their archiving to tapes for the LTS, adhering to the prerequisites set by the Rectorate (UNIRIS) and DCSR. For more details, see archiving prerequisites.
- README files are prepared automatically for sharing data on generalist external repositories (e.g., Zenodo or Dryad) during publication processes.
- Metadata is tailored for sharing on specialized repositories (e.g. OMICS data or other domain-specific datasets).
Characteristics of a readme file
A README file is a crucial component of dataset documentation, providing essential information to ensure that the data can be accurately interpreted and utilized by others, as well as by yourself in the future. It serves to enhance the usability, reproducibility, and transparency of your dataset.
Key Elements to Include in a Data README File:
General Information:
Dataset Title: Provide a clear and descriptive title for your dataset.
Author Information: List the names, affiliations, and contact details of the principal investigator and any co-investigators.
Date of Data Collection: Specify the dates when the data was collected.
Geographical Location: If applicable, mention where the data was collected.
Keywords: Include relevant keywords to describe the data’s subject matter.
Data and File Overview:
File Descriptions: Provide a brief description of each file, including its format and purpose.
File Structure: Explain the organization of the files and any relationships between them.
File Naming Conventions: Describe the naming conventions used for files and directories.
Methodological Information:
Data Collection Methods: Detail the procedures and instruments used to collect the data.
Data Processing: Explain any processing or transformation applied to the data.
Quality Assurance: Describe steps taken to ensure data quality and integrity.
Data-Specific Information:
Variable Definitions: Define all variables, including units of measurement and possible codes.
Missing Data: Specify how missing data is represented in the dataset.
Data Formats: Indicate any specialized formats or abbreviations used.
Sharing and Access Information:
Licenses or Restrictions: State any licenses or
restrictions associated with the data.
Related Publications: Provide references to publications that use or are related to the data.
Citation Information: Offer a recommended citation for the dataset.
Best Practices:
File Format: Write the README as a plain text file (e.g., README.txt) to ensure accessibility and longevity.
Standardized Formatting: Use consistent formatting and terminology throughout the README.
Clarity and Detail: Provide sufficient detail to allow others to understand and use the data without additional assistance.
For comprehensive guidance and templates, consider consulting DSBU website.
Structured Data Management:
- The platform provides guidance on file format standards, disciplinary metadata, and recommends relevant FAIR repositories for data sharing.
- DataSquid helps you structure your data on the NAS -DCSR and ensures easy access to all related folders and documentation.
Details
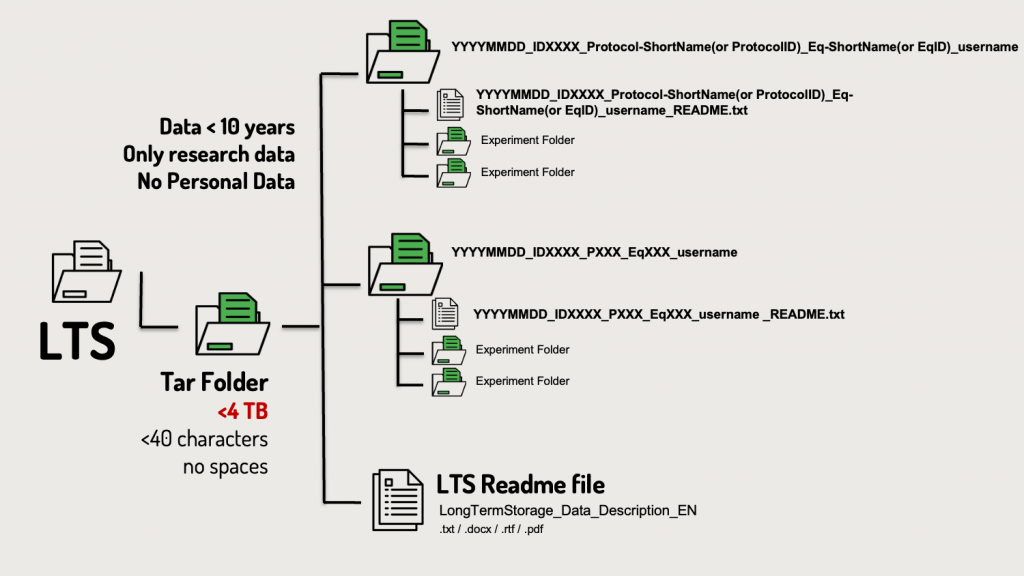
Folder Structure
The data is organized in a structured folder hierarchy to ensure clarity and consistency. Each folder represents a specific experiment and contains relevant subfolders and files.
Main Organizational Structure
• Subfolders: Each folder corresponds to a single experiment and contains:
o Data files/folders: The acquisition or analysis data.
o README files: These txt documents explain the content and structure of the subfolder.
Subfolder Naming Convention
Each subfolder is named to represent a single experiment using the following structure:
YYYYMMDD_IDXXXX_Protocol-ShortName(or ProtocolID)_Eq-ShortName(EqID)_username
Explanation of Subfolder Naming Components:
- YYYYMMDD:
The date when the data was collected, formatted as YYYYMMDD (e.g., 20231125 for November 25, 2023). - IDXXXX:
A unique identifier representing the acquisition or analysis, either:
o ID (for acquisition) - Protocol-ShortName (or ProtocolID):
The internal name or ID of the protocol used during data acquisition (e.g., P001). - Eq-ShortName (or EqID):
The name or ID of the equipment used for the experiment (e.g., SP5confocal). - username:
The UNIL username of the experimenter responsible for the data (e.g., clebrand).
The README file naming follows the rule:
Subfolder Name + ‘_README.txt’.
For example, if the subfolder is named 20231125_ID001_P001_SP5confocal_clebrand, the README file would be named: 20231125_ID001_P001_SP5confocal_clebrand_README.txt.
Effortless Project Integration:
- The DSBU automatically imports and updates projects from the NAS-DCSR into DataSquid weekly, requiring no action from researchers.
- The Principal Investigator (PI) only needs to validate new group members when they join a project on DataSquid.
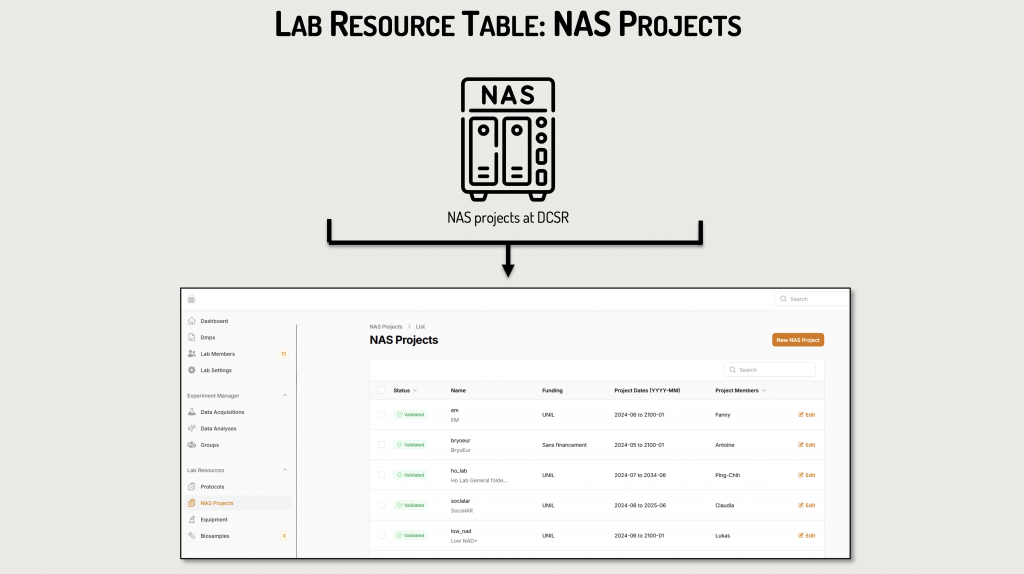
Preloaded Resources:
The DSBU preloads critical resources into the DataSquid ressources tables, including:
- Data category and Data Sub-category: consult our FBM Data classification
- Equipment and software for data acquisition and analyses deployed by FBM platforms
- Biosamples and via uploaded tables generated by other LIMS (GTF, PAF, EMF)
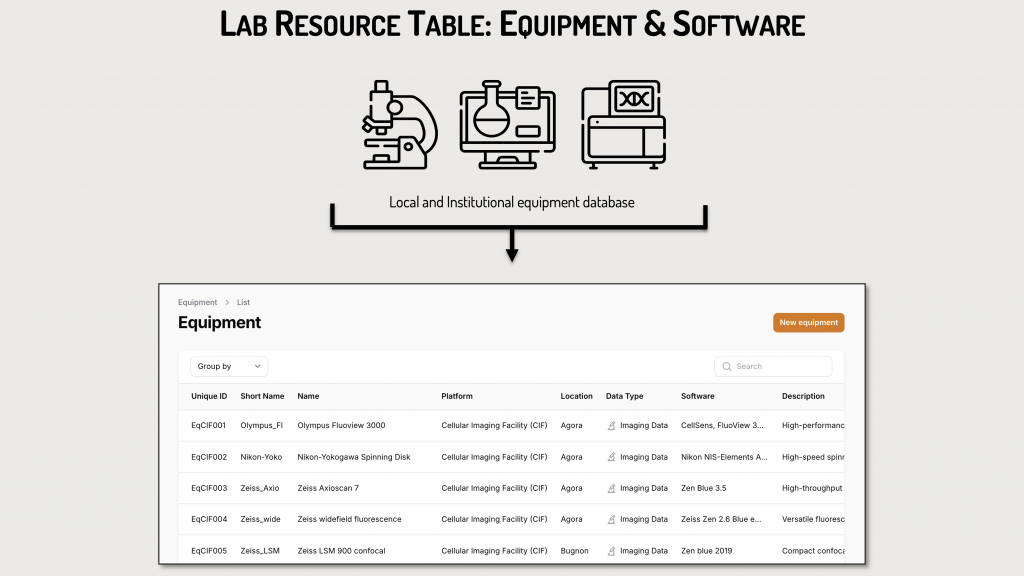
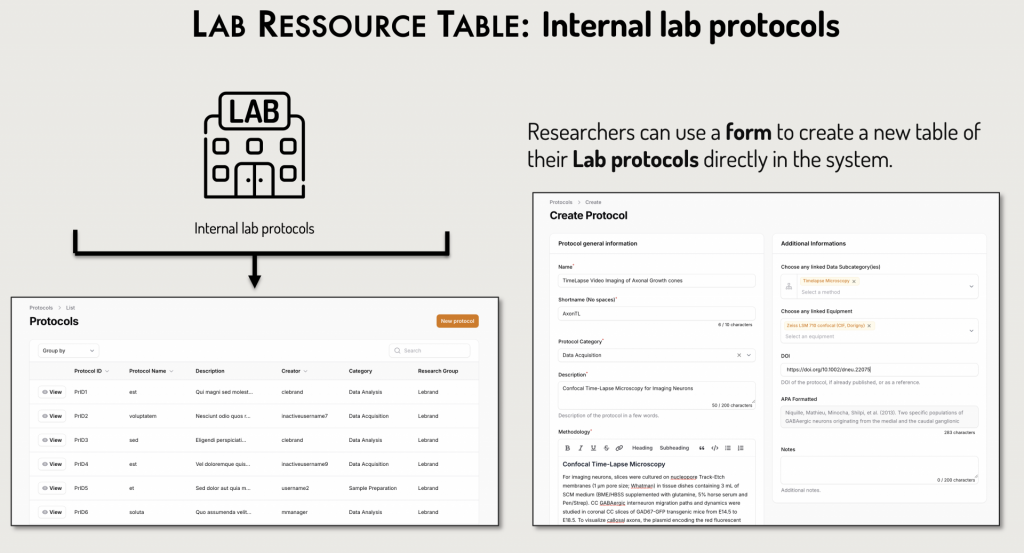
Custom Additions by Researchers:
Researchers can proactively add their own resources:
- Equipment specific to their laboratory or department using an intuitive form.
- Lab Biosamples via uploaded tables (e.g., Pyrat) or by creating a new table directly in DataSquid.
- Lab protocols, which can be added through a form or as new tables.
Efficient Workflow Integration:
- Intuitive Workflow: DataSquid provides a user-friendly process for managing research documentation for each individual experiment.
- Resource Integration: Once resource information is entered into the resource tables, the platform automatically integrates it into the workflow.
- Experiment Documentation: Streamlines the creation of detailed documentation for every new experiment or acquisition in the lab.
- Best Practices Compliance: Ensures that all activities are fully documented and aligned with research best practices.
- Enhanced Efficiency: Promotes consistent and efficient research data management across the lab.
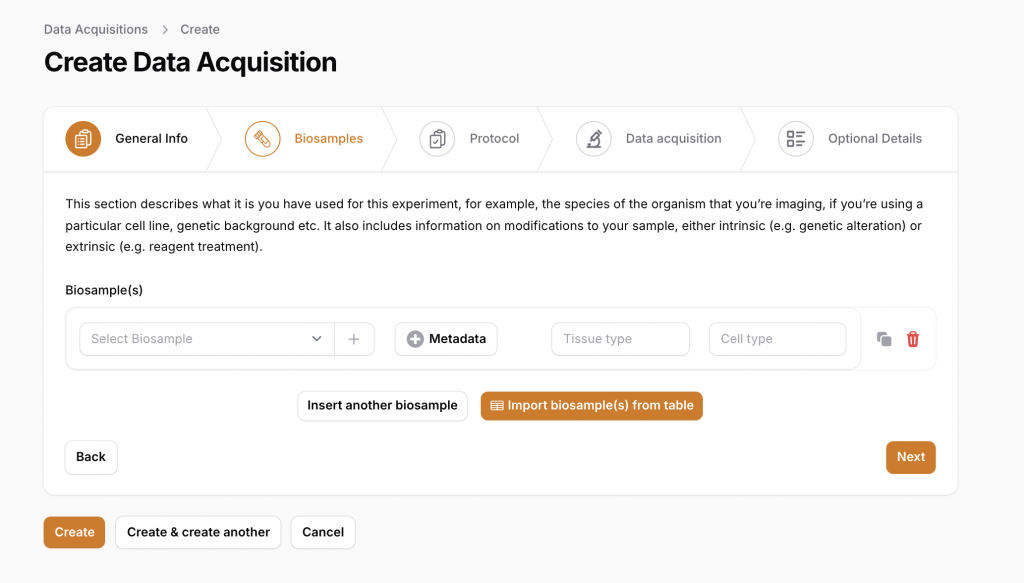
Proactive and Retrospective Documentation Creation:
DataSquid is user-friendly, enabling:
- Proactive documentation: README files and metadata are automatically created for new acquisitions or analyses.
- Retrospective documentation: If experiments were conducted before information was added to DataSquid, the tool generates a simplified README to document the past experiments efficiently.

With its intuitive design and automation features, DataSquid significantly reduces administrative tasks, ensures adherence to data standards, and facilitates compliance with FAIR principles for data sharing.
