2025
Sex-Specific Effects of Hypocretin Receptor Signaling in CRF Neurons on Alcohol Drinking, Anxiety, and BNST Neuronal Excitability
Abstract
Background
Alcohol use disorder (AUD) is characterized by compulsive alcohol consumption and negative emotional states during withdrawal, often perpetuating a cycle of addiction through arousal dysfunction. The hypocretin/orexin (Hcrt) neuropeptide system is a key regulator of arousal that is implicated in these processes, particularly in its interactions with corticotropin-releasing factor (CRF) neurons within the bed nucleus of the stria terminalis (BNST).
Methods
Using CRF-specific genetic deletion of HcrtR1 and/or HcrtR2 receptors in mice, combined with behavioral and electrophysiological approaches, we investigated the role of Hcrt receptor signaling in CRF neurons in modulating alcohol intake, anxiety behaviors, and BNST excitability, with a focus on sex-specific differences.
Results
We found that deletion of HcrtR1 significantly reduced alcohol intake, with sex-specific effects on BNST excitability and synaptic drive. CRF-specific HcrtR2 deletion, while not affecting alcohol consumption, decreased baseline anxiety-like behaviors in males relative to females. Moreover, the double deletion of both Hcrt receptors from CRF neurons led to reduced alcohol drinking in males (while tending to increase alcohol drinking in females) and dampened anxiety behaviors and BNST excitability in both sexes during protracted withdrawal.
Conclusions
These findings suggest that Hcrt signaling in CRF neurons plays a critical role in the persistence of excessive alcohol consumption and the development of negative affective states, with distinct contributions from HcrtR1 and HcrtR2. The observed sex-specific differences underscore the need for tailored therapeutic approaches targeting the Hcrt system in the treatment of AUD.
- Yihe Ma, Haniyyah Sardar, Max E. Benabou, Angeline C. Yu, Allison R. Morningstar, R.Nicolas Fajardo, Isaac F. Kandil, Ethan T. Rogers, Anne Vassalli, Julie A. Kauer, William J. Giardino
- Biological Psychiatry Global Open Science, 2025 Sep 20, 100617, ISSN 2667-1743, DOI: 10.1016/j.bpsgos.2025.100617.
- https://www.sciencedirect.com/science/article/pii/S2667174325001715
Contrasting contribution of resident and repopulated brain macrophages in sustaining sleep-wake circuitry
Abstract
Sleep is a complex behavior regulated by various brain cell types. However, the roles of brain resident macrophages, including microglia and CNS-associated macrophages (CAMs), particularly those derived postnatally, in sleep regulation remain poorly understood. Here, we investigated the effects of resident (embryo-derived) and repopulated (postnatally derived) brain-resident macrophages on the regulation of vigilance states in mice. We found that depletion in resident brain macrophages caused increased sleep in the active period, but reduced its quality, reflected in reduced power of brain sleep oscillations. This was observed both for the Non-REM and REM sleep stages. Subsequent repopulation by postnatal brain macrophages resulted in altered, but not fully restored, sleep-wake patterns and additionally induced sleep fragmentation. Furthermore, brain macrophage depletion caused excitatory inhibitory synaptic imbalance, which was resistant to repopulation, and led to increased inhibitory synapses. At the metabolite level, the distinct metabolite profile induced by brain macrophage depletion largely returned to normal after repopulation. Our findings suggest a so far largely unknown interaction between brain-resident macrophages and sleep and highlight functional differences between resident and postnatally-derived repopulated brain macrophages, paving the way to future exploration of the role of brain macrophages of different origin in sleep disorders and synaptic connectivity.
- Seifinejad A, Bandarabadi M, Haddar M, Wundt S, Tafti M, Vassalli A, Khani A, Monaco G.
- Commun Biol. 2025 Sep 9;8(1):1339. DOI: 10.1038/s42003-025-08781-7.PMID: 40925913 Free PMC article.
- https://www.nature.com/articles/s42003-025-08781-7
Divergent modulation of dopaminergic neurons by hypocretin/orexin receptors-1 and -2 shapes dopaminergic cell activity and socio-emotional behavior
Stamatina Tzanoulinou, Simone Astori, Laura Clara Grandi, Francesca Gullo, Richie Kalusivikako, Simran Rai, Mehdi Tafti, Andrea Becchetti, and Anne Vassalli
Abstract
Many neuropsychiatric disorders involve dysregulation of the dopaminergic (DA) input to the forebrain. Of particular relevance are DA projections originating from the midbrain ventral tegmental area (VTA). A key neuromodulatory influence onto DAVTA neurons arises from the dense axonal projections emanating from lateral hypothalamic area hypocretin/orexin (OX) neurons. Despite being a major input, the differential action of orexin peptides A and B (OXA and OXB) on orexin receptors 1 and 2 (OX1R and OX2R) in DA cells is poorly characterized. We thus genetically engineered mice whose DA neurons are selectively unresponsive to OX input via OX1R (DAOx1R-KO mice) or OX2R (DAOx2R-KO mice) and compared their behavior and DA cell electrophysiology to genetic controls. We previously showed a profound functional divergence between OX1R- and OX2R-mediated modulation of DA neurons in controlling vigilance states, brain oscillations and cognitive behavior. Inactivation of OX2R, but not OX1R, in DA neurons dramatically increased time spent in EEG theta-rich wakefulness, improved reward-driven learning and attentional skills, while it impaired inhibitory control. Here, we interrogate DAOx1R-KO and DAOx2R-KO mice in further behavioral domains. We show that mice with DA-specific OX1R loss exhibit hyperactivity, or anxiety-like responding, in context-dependent manners. OX2R loss in contrast decreases sociability and aversion-driven learning. We next investigate the underlying electrophysiological substrates and uncover previously unrecognized effects of OX peptides on DAVTA cell responses. In WT and control mice, we show that while OXA enhances, OXB diminishes DAVTA neuronal excitability. OX1R-deficient DA cells lose OXA responding and OX2R-deficient DA cells lose OXB responding. We altogether evidence strikingly distinct functions of OX1 vs OX2R signaling in modulating the intrinsic excitability of DAVTA neurons and influencing DA-related behaviors. Our data position OX→DA neurotransmission via OX1 or OX2R as relevant to endophenotypes observed in the context of disorders such as obsessive-compulsive, attention-deficit/hyperactivity, and autism spectrum disorders.

Graphical Abstract: Summary depiction of divergent neuromodulation of dopaminergic neurons by hypocretin/orexin receptors-1 and -2 as reflected at electrophysiological and behavioral levels
https://www.biorxiv.org/content/10.1101/2025.02.14.638329v2
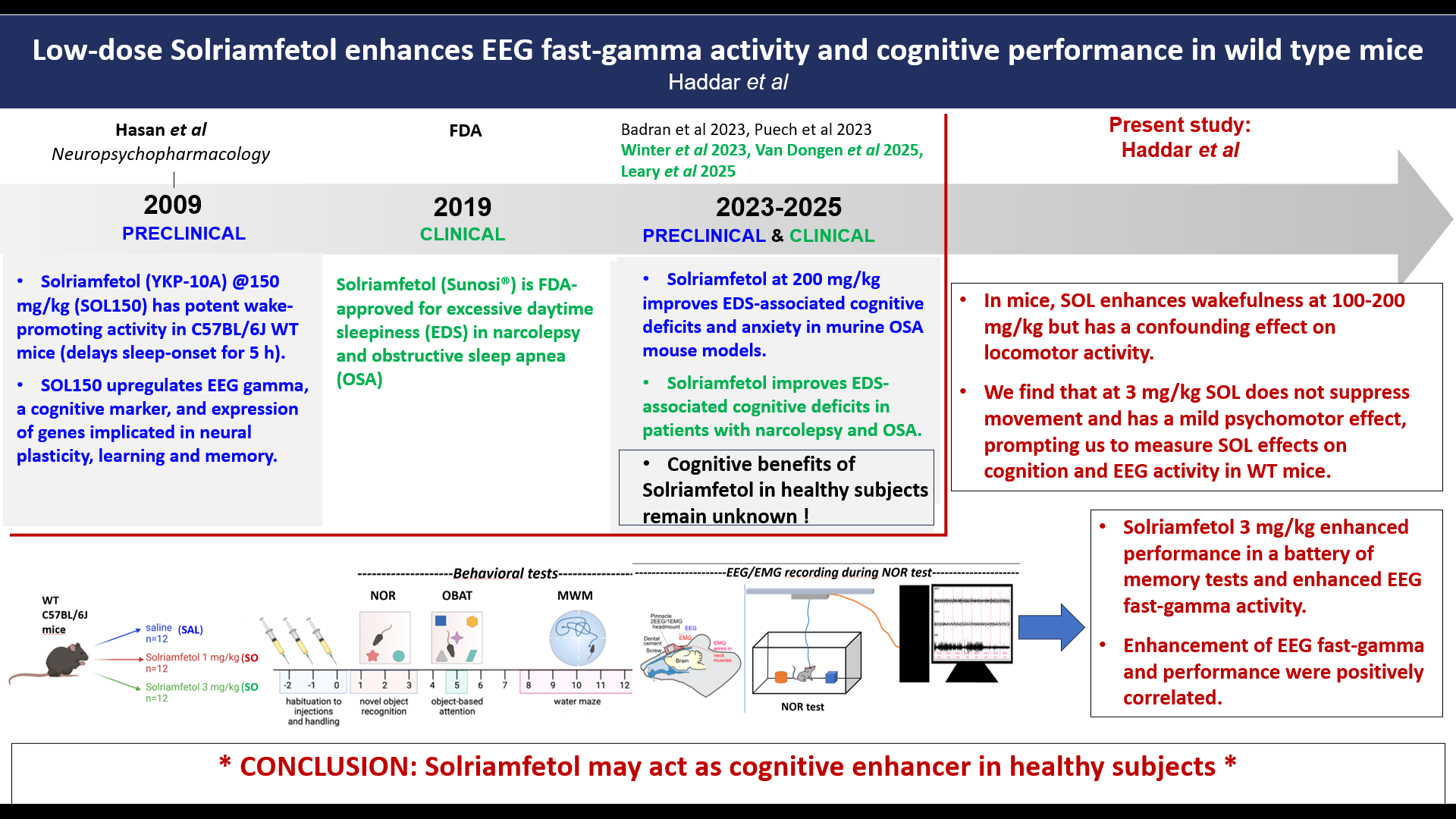
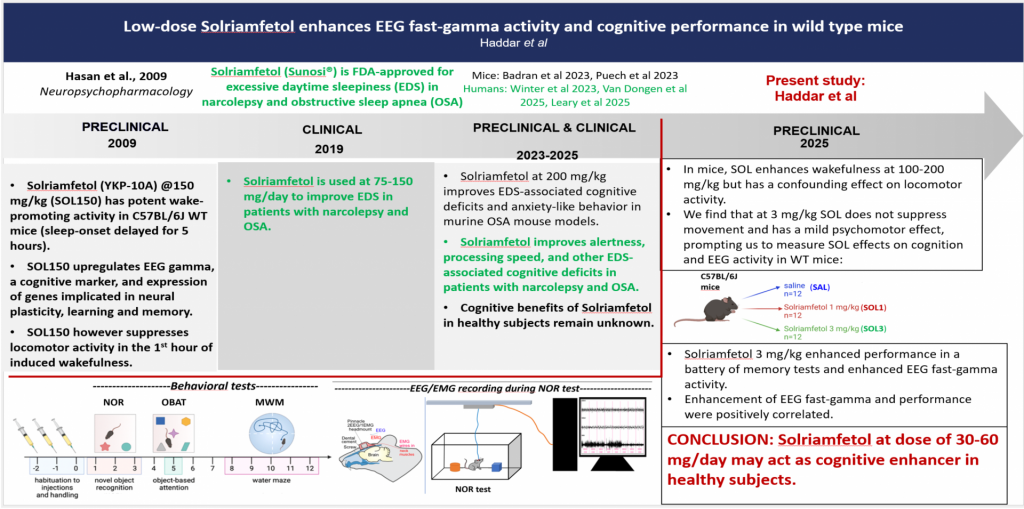
Solriamfetol enhances alertness and cognitive performance in mice
Meriem Haddar, Stamatina Tzanoulinou, Li Yuan Chen, Mehdi Tafti, and Anne Vassalli
Abstract
Solriamfetol [(R)-2-amino-3-phenylpropylcarbamate hydrochloride], a phenylalanine derivative initially developed as potential antidepressant, was shown by our group in 2009 to have potent, dose-dependent wake-promoting activity in mice. Solriamfetol (Sunosi®) is used since 2019 to counteract excessive daytime sleepiness (EDS) in patients with narcolepsy and obstructive sleep apnea (OSA). It has several advantages over other stimulants, notably that it is not associated with strong psychomotor activity, does not induce behavioral stereotypies and anxiety-related behaviors, in contrast to amphetamines and modafinil. Its mode-of-action remains incompletely solved. It was reported to act as dual dopamine-and-noradrenaline-reuptake-inhibitor (DNRI), and more recently, to have TAAR1 agonist activity. In our early mouse study, we showed that, at 150 mg/kg, Solriamfetol induces a state of wakefulness featuring a dramatic upregulation of EEG gamma activity, a cognitive biomarker, and the expression of genes implicated in neural plasticity, learning and memory. Besides being prescribed to treat EDS, central nervous system stimulants are commonly used as ‘smart drugs’ to enhance cognition in normal individuals. Therefore, based on Solriamfetol’s ability to potently induce wakefulness, EEG and molecular markers of learning and memory, we aimed to determine whether it could improve the cognitive performance of wild-type mice. Because at doses of 50-150 mg/kg, Solriamfetol induced an alert waking state associated with low mobility, thus precluding behavioral testing, we used lower doses (1-3 mg/kg) to assess cognition in a battery of tests evaluating short- or long-term memory and spatial navigation. We found that compared to saline, Solriamfetol 3 mg/kg consistently improves sustained attention for a novel object, as well as spatial memory. Next, to determine the brain activity correlates of enhanced cognition, we performed EEG/EMG recording while the mice performed the novel object recognition (NOR) task. Power spectral density (PSD) analysis revealed that 3 mg/kg Solriamfetol reduced EEG delta (an index of sleepiness) during exposure to a novel context, and enhanced EEG fast-gamma (an index of mental concentration) during execution of the NOR task. Taken together, our data demonstrate that low-dose Solriamfetol improves memory and attentional performance in wild-type mice.
https://www.biorxiv.org/content/10.1101/2025.01.02.631072v1
2024
Sex-Specific Roles of Hypocretin Receptor Signaling in CRF Neurons on Alcohol Drinking, Anxiety, and BNST Neuronal Excitability.
Ma Y, Sardar H, Benabou ME, Yu AC, Morningstar AR, Fajardo RN, Kandil IF, Rogers ET, Vassalli A, Kauer JA, Giardino WJ.
doi:10.1101/2024.09.07.609774.PMID: 39282398 (Accepted in Biological Psychiatry Global Open Science, 2025 Sep 20)
Summary
It is suggested that Hcrt signaling in CRF neurons plays a critical role in the persistence of excessive alcohol consumption and the development of negative affective states, with distinct contributions from HcrtR1 and HcrtR2.
Abstract
Alcohol use disorder (AUD) is characterized by compulsive alcohol consumption and negative emotional states during withdrawal, often perpetuating a cycle of addiction through arousal dysfunction. The hypocretin/orexin (Hcrt) neuropeptide system, a key regulator of arousal, has been implicated in these processes, particularly in its interactions with corticotropin-releasing factor (CRF) neurons within the bed nucleus of the stria terminalis (BNST). We investigated the role of Hcrt receptor signaling in CRF neurons in modulating alcohol intake, anxiety behaviors, and BNST excitability, with a focus on sex-specific differences. Using CRF-specific genetic deletion of HcrtR1 and/or HcrtR2 receptors in mice, we found that deletion of HcrtR1 significantly reduced alcohol intake, with sex-specific effects on BNST excitability. CRF-specific HcrtR2 deletion, while not affecting alcohol consumption, decreased baseline anxiety-like behaviors in males relative to females. Moreover, the double deletion of both Hcrt receptors from CRF neurons led to reduced alcohol drinking in males and dampened anxiety behaviors and BNST excitability in both sexes during protracted withdrawal. These findings suggest that Hcrt signaling in CRF neurons plays a critical role in the persistence of excessive alcohol consumption and the development of negative affective states, with distinct contributions from HcrtR1 and HcrtR2. The observed sex-specific differences underscore the need for tailored therapeutic approaches targeting the Hcrt system in the treatment of AUD.
Inactivation of hypocretin receptor-2 signaling in dopaminergic neurons induces hyperarousal and enhanced cognition but impaired inhibitory control.
Bandarabadi M, Li S, Aeschlimann L, Colombo G, Tzanoulinou S, Tafti M, Becchetti A, Boutrel B, and Vassalli A.
Mol Psychiatry. 2024 Feb;29(2):327-341.
doi:10.1038/s41380-023-02329-z.
Hypocretin/Orexin (HCRT/OX) and dopamine (DA) are both key effectors of salience processing, reward and stress-related behaviors and motivational states, yet their respective roles and interactions are poorly delineated. We inactivated HCRT-to-DA connectivity by genetic disruption of Hypocretin receptor-1 (Hcrtr1), Hypocretin receptor-2 (Hcrtr2), or both receptors (Hcrtr1&2) in DA neurons and analyzed the consequences on vigilance states, brain oscillations and cognitive performance in freely behaving mice. Unexpectedly, loss of Hcrtr2, but not Hcrtr1 or Hcrtr1&2, induced a dramatic increase in theta (7-11 Hz) electroencephalographic (EEG) activity in both wakefulness and rapid-eye-movement sleep (REMS). DAHcrtr2-deficient mice spent more time in an active (or theta activity-enriched) substate of wakefulness, and exhibited prolonged REMS. Additionally, both wake and REMS displayed enhanced theta-gamma phase-amplitude coupling. The baseline waking EEG of DAHcrtr2-deficient mice exhibited diminished infra-theta, but increased theta power, two hallmarks of EEG hyperarousal, that were however uncoupled from locomotor activity. Upon exposure to novel, either rewarding or stress-inducing environments, DAHcrtr2-deficient mice featured more pronounced waking theta and fast-gamma (52-80 Hz) EEG activity surges compared to littermate controls, further suggesting increased alertness. Cognitive performance was evaluated in an operant conditioning paradigm, which revealed that DAHcrtr2-ablated mice manifest faster task acquisition and higher choice accuracy under increasingly demanding task contingencies. However, the mice concurrently displayed maladaptive patterns of reward-seeking, with behavioral indices of enhanced impulsivity and compulsivity. None of the EEG changes observed in DAHcrtr2-deficient mice were seen in DAHcrtr1-ablated mice, which tended to show opposite EEG phenotypes. Our findings establish a clear genetically-defined link between monosynaptic HCRT-to-DA neurotransmission and theta oscillations, with a differential and novel role of HCRTR2 in theta-gamma cross-frequency coupling, attentional processes, and executive functions, relevant to disorders including narcolepsy, attention-deficit/hyperactivity disorder, and Parkinson’s disease.

https://www.nature.com/articles/s41380-023-02329-z
Contrasting contribution of embryo- and postnatally-derived brain-resident macrophages in sustaining sleep-wake circuitryAli Seifinejad, Mojtaba Bandarabadi, Meriem Haddar, Saskia Wundt, Mehdi Tafti, Anne Vassalli, Abbas Khani, Gianni Monaco
bioRxiv 2024.01.22.576653; doi: https://doi.org/10.1101/2024.01.22.576653
Abstract
Sleep is a complex behavior regulated by a variety of brain cell types. The roles of brain-resident macrophages, such as microglia and CNS-associated macrophages (CAMs), including those derived postnatally, are not well defined. Here, we investigated the reciprocal interaction of brain-resident macrophages and sleep using multimodal high-throughput transcriptional, electrophysiological and metabolomic profiling in mice. We found that sleep deprivation caused profound transcriptional changes in microglia and CAMs, which were intensified by impaired sleep regulation in the absence of the important sleep-regulatory neuropeptide hypocretin/orexin (HCRT). Depletion in embryonically-derived brain macrophages caused increased sleep in the active period, but reduced its quality, reflected in reduced power of brain sleep oscillations. This was observed both for the Non-REM and REM sleep stages. Subsequent repopulation by postnatal brain macrophages unexpectedly failed to reestablish normal sleep-wake patterns and additionally induced sleep fragmentation. Furthermore, brain macrophage depletion caused excitatory-inhibitory synaptic imbalance, which was resistant to repopulation, and led to increased inhibitory synapses. At the metabolite level, the distinct metabolite profile induced by brain macrophage depletion largely returned to normal after repopulation. Our findings suggest a so far largely unknown interaction between brain-resident macrophages and sleep and emphasizes striking functional differences between embryonic and postnatally-derived brain macrophages, paving the way to future exploration of the role of brain macrophages of different origin in sleep disorders and synaptic connectivity.
2023
Epigenetic silencing of selected hypothalamic neuropeptides in narcolepsy with cataplexy.
Seifinejad A, Ramosaj M, Shan L, Li S, Possovre ML, Pfister C, Fronczek R, Garrett-Sinha LA, Frieser D, Honda M, Arribat Y, Grepper D, Amati F, Picot M, Agnoletto A, Iseli C, Chartrel N, Liblau R, Lammers GJ, Vassalli A, Tafti M.
Proc Natl Acad Sci U S A. 2023 May 9;120(19):e2220911120. doi: 10.1073/pnas.2220911120. Epub 2023 May 1.PMID: 37126681
Summary
It is demonstrated that a substantial number of HCRT neurons are present in the brain of patients, but HCRT gene is silenced by methylation, and possibilities exist to reactivate them to treat or even cure narcolepsy.
Abstract
Narcolepsy with cataplexy is characterized by excessive daytime sleepiness and sudden loss of muscle tone (cataplexy) triggered by strong emotions. The best marker of the disease is low/absent hypocretin (HCRT), a hypothalamic neuropeptide. An autoimmune attack targeting these neurons is believed to cause HCRT deficiency, despite any direct evidence. We demonstrate that a substantial number of HCRT neurons are present in the brain of patients, but HCRT gene is silenced by methylation. The epigenetic silencing is not restricted to HCRT but also CRH and PDYN genes are methylated in the hypothalamus of patients. Our results strongly suggest that HCRT and CRH neurons are not destroyed but epigenetically deactivated, and possibilities exist to reactivate them to treat or even cure narcolepsy.
Hypothalamic control of noradrenergic neurons stabilizes sleep, Gianandrea Broglia, Giorgio Corsi, Pierre-Hugues Prouvot Bouvier, Anne Vassalli, Mehdi Tafti, Mojtaba Bandarabadi.
bioRxiv. 2023. https://doi.org/10.1101/2023.10.22.563502
Abstract
Hypocretin/orexin neurons are essential to stabilize sleep, but the underlying mechanisms remain elusive. We report that hypocretin neurons of the perifornical hypothalamus are highly active during rapid eye movement sleep and show state-specific correlation with noradrenergic neurons. Deletion of hypocretin gene significantly increased periodic reactivations of locus coeruleus noradrenergic neurons during sleep and dysregulated their activity across transitions, suggesting a role for hypocretin neurons in mediating neuromodulation to stabilize sleep.
2022
2022
Orexin action on the dopaminergic system modulates theta during REM sleep and wakefulness
Mojtaba Bandarabadi, Sha Li, Mehdi Tafti, Giulia Colombo, Andrea Becchetti, Anne Vassalli
bioRxiv 2022.01.30.478401; doi: https://doi.org/10.1101/2022.01.30.478401
Abstract
Both dopaminergic (DA) and orexinergic (OX) systems establish brain-wide neuromodulatory circuits that profoundly influence brain states and behavioral outputs. To unravel their interactions, we inactivated OX-to-DA neurotransmission by selective disruption of HcrtR1/OxR1, or HcrtR2/OxR2, or both receptors, in DA neurons. Chronic loss of OXR2 in DA neurons (OxR2Dat-CKO mice) dramatically increased electrocorticographic (EcoG) theta rhythms in wakefulness and REM sleep. Episode duration and total times spent in ‘active’ wakefulness and REMS were prolonged, and theta/fast-gamma wave coupling was enhanced in both states. Increased theta in OxR2DatCKO mice baseline wake was accompanied by diminished infra-theta and increased fast-gamma activities, i.e. the mice exhibited signs of constitutive electrocortical hyperarousal, albeit uncoupled with locomotor activity. These effects were not seen in OxR1-ablated dopaminergic mutants, which tended to show opposite phenotypes, resembling those caused by the loss of both receptors. Our data establish a clear, genetically-defined link between monosynaptic orexin-to-dopaminergic connectivity and the power of theta oscillations, with a differential role of OXR2 in cross-frequency wave coupling and attentional processes.
Neurobiology of cataplexy.
Seifinejad A, Vassalli A, Tafti M.
Sleep Med Rev. 2021 Dec;60:101546. doi: 10.1016/j.smrv.2021.101546. Epub 2021 Sep 20.PMID: 34607185
Abstract
Cataplexy is the pathognomonic and the most striking symptom of narcolepsy. It has originally been, and still is now, widely considered as an abnormal manifestation of rapid eye movement (REM) sleep during wakefulness due to the typical muscle atonia. The neurocircuits of cataplexy, originally confined to the brainstem as those of REM sleep atonia, now include the hypothalamus, dorsal raphe (DR), amygdala and frontal cortex, and its neurochemistry originally focused on catecholamines and acetylcholine now extend to hypocretin (HCRT) and other neuromodulators. Here, we review the neuroanatomy and neurochemistry of cataplexy and propose that cataplexy is a distinct brain state that, despite similarities with REM sleep, involves cataplexy-specific features.
2020
Hypocretinergic interactions with the serotonergic system regulate REM sleep and cataplexy.
Seifinejad A, Li S, Possovre ML, Vassalli A, Tafti M.
Nat Commun. 2020 Nov 27;11(1):6034. doi: 10.1038/s41467-020-19862-y.PMID: 33247179
Summary
It is shown that deleting the serotonin transporter gene in hypocretin knockout mice suppresses cataplexy while dramatically increasing REM sleep, indicating that these are two different states but are both regulated by hypocrretinergic input to serotonergic neurons.
Abstract
Loss of muscle tone triggered by emotions is called cataplexy and is the pathognomonic symptom of narcolepsy, which is caused by hypocretin deficiency. Cataplexy is classically considered to be an abnormal manifestation of REM sleep and is treated by selective serotonin (5HT) reuptake inhibitors. Here we show that deleting the 5HT transporter in hypocretin knockout mice suppressed cataplexy while dramatically increasing REM sleep. Additionally, double knockout mice showed a significant deficit in the buildup of sleep need. Deleting one allele of the 5HT transporter in hypocretin knockout mice strongly increased EEG theta power during REM sleep and theta and gamma powers during wakefulness. Deleting hypocretin receptors in the dorsal raphe neurons of adult mice did not induce cataplexy but consolidated REM sleep. Our results indicate that cataplexy and REM sleep are regulated by different mechanisms and both states and sleep need are regulated by the hypocretinergic input into 5HT neurons. Narcolepsy is characterized by a sudden loss of muscle tone (cataplexy) similar to REM sleep and is caused by hypocretin deficiency. Here, the authors show that deleting the serotonin transporter gene in hypocretin knockout mice suppresses cataplexy while dramatically increasing REM sleep, indicating that these are two different states but are both regulated by hypocretinergic input to serotonergic neurons.
Sleep as a default state of cortical and subcortical networks
Mojtaba Bandarabadi; Anne Vassalli; Mehdi Tafti
Current Opinion in Physiology
2020-06 | Journal article
DOI: 10.1016/j.cophys.2019.12.004
Molecular codes and in vitro generation of hypocretin and melanin concentrating hormone neurons.
Seifinejad A, Li S, Mikhail C, Vassalli A, Pradervand S, Arribat Y, Pezeshgi Modarres H, Allen B, John RM, Amati F, Tafti M.
Proc Natl Acad Sci U S A. 2019 Aug 20;116(34):17061-17070. doi: 10.1073/pnas.1902148116. Epub 2019 Aug 2.PMID: 31375626
Summary
It is discovered that partial removal of PEG3 in mice significantly reduces the number of HCRT and melanin concentrating hormone neurons, and its down-regulation in zebrafish completely abolishes their expression, resulting in a 2-fold increase in sleep amount.
Abstract
Hypocretin (HCRT) and melanin concentrating hormone are brain neuropeptides involved in multiple functions, including sleep and metabolism. Loss of HCRT causes the sleep disorder narcolepsy. To understand how these neuropeptides are produced and contribute to diverse functions in health and disease, we purified their cells from mouse embryonic brains and established their molecular machinery. We discovered that partial removal of PEG3 (a transcription factor) in mice significantly reduces the number of HCRT and melanin concentrating hormone neurons, and its down-regulation in zebrafish completely abolishes their expression. We used our molecular data to produce these neurons in vitro from mouse fibroblasts, a technique that can be applied to cells from narcolepsy patients to generate an in vitro cell-based model. Hypocretin/orexin (HCRT) and melanin concentrating hormone (MCH) neuropeptides are exclusively produced by the lateral hypothalamus and play important roles in sleep, metabolism, reward, and motivation. Loss of HCRT (ligands or receptors) causes the sleep disorder narcolepsy with cataplexy in humans and in animal models. How these neuropeptides are produced and involved in diverse functions remain unknown. Here, we developed methods to sort and purify HCRT and MCH neurons from the mouse late embryonic hypothalamus. RNA sequencing revealed key factors of fate determination for HCRT (Peg3, Ahr1, Six6, Nr2f2, and Prrx1) and MCH (Lmx1, Gbx2, and Peg3) neurons. Loss of Peg3 in mice significantly reduces HCRT and MCH cell numbers, while knock-down of a Peg3 ortholog in zebrafish completely abolishes their expression, resulting in a 2-fold increase in sleep amount. We also found that loss of HCRT neurons in Hcrt-ataxin-3 mice results in a specific 50% decrease in another orexigenic neuropeptide, QRFP, that might explain the metabolic syndrome in narcolepsy. The transcriptome results were used to develop protocols for the production of HCRT and MCH neurons from induced pluripotent stem cells and ascorbic acid was found necessary for HCRT and BMP7 for MCH cell differentiation. Our results provide a platform to understand the development and expression of HCRT and MCH and their multiple functions in health and disease.
Sleep modulates haematopoiesis and protects against atherosclerosis.
McAlpine CS, Kiss MG, Rattik S, He S, Vassalli A, Valet C, Anzai A, Chan CT, Mindur JE, Kahles F, Poller WC, Frodermann V, Fenn AM, Gregory AF, Halle L, Iwamoto Y, Hoyer FF, Binder CJ, Libby P, Tafti M, Scammell TE, Nahrendorf M, Swirski FK.
Nature. 2019 Feb;566(7744):383-387. doi: 10.1038/s41586-019-0948-2. Epub 2019 Feb 13.PMID: 30760925
Summary
The fragmentation of sleep in Apoe−/− mice induces monocytosis and accelerated atherosclerosis due to a reduction in hypocretin that otherwise restricts bone marrow CSF1 availability, and this results identify a neuro-immune axis that links sleep to haematopoiesis and Atherosclerosis.
Abstract
Sleep is integral to life. Although insufficient or disrupted sleep increases the risk of multiple pathological conditions, including cardiovascular disease, we know little about the cellular and molecular mechanisms by which sleep maintains cardiovascular health. Here we report that sleep regulates haematopoiesis and protects against atherosclerosis in mice. We show that mice subjected to sleep fragmentation produce more Ly-6C high monocytes, develop larger atherosclerotic lesions and produce less hypocretin—a stimulatory and wake-promoting neuropeptide—in the lateral hypothalamus. Hypocretin controls myelopoiesis by restricting the production of CSF1 by hypocretin-receptor-expressing pre-neutrophils in the bone marrow. Whereas hypocretin-null and haematopoietic hypocretin-receptor-null mice develop monocytosis and accelerated atherosclerosis, sleep-fragmented mice with either haematopoietic CSF1 deficiency or hypocretin supplementation have reduced numbers of circulating monocytes and smaller atherosclerotic lesions. Together, these results identify a neuro-immune axis that links sleep to haematopoiesis and atherosclerosis. The fragmentation of sleep in Apoe−/− mice induces monocytosis and accelerated atherosclerosis due to a reduction in hypocretin that otherwise restricts bone marrow CSF1 availability.
Bidirectional and context-dependent changes in theta and gamma oscillatory brain activity in noradrenergic cell-specific Hypocretin/Orexin receptor 1-KO mice.
Li S, Franken P, Vassalli A.
Sci Rep. 2018 Oct 19;8(1):15474. doi: 10.1038/s41598-018-33069-8.PMID: 30341359
Summary
HCRT-to-NA signalling may fine-tune arousal, up in alarming conditions, and down during self-motivated, goal-driven behaviors, as well as regulates the slow-δ rebound characterizing sleep after stress-associated arousal.
Abstract
Noradrenaline (NA) and hypocretins/orexins (HCRT), and their receptors, dynamically modulate the circuits that configure behavioral states, and their associated oscillatory activities. Salient stimuli activate spiking of locus coeruleus noradrenergic (NALC) cells, inducing NA release and brain-wide noradrenergic signalling, thus resetting network activity, and mediating an orienting response. Hypothalamic HCRT neurons provide one of the densest input to NALC cells. To functionally address the HCRT-to-NA connection, we selectively disrupted the Hcrtr1 gene in NA neurons, and analyzed resulting (Hcrtr1Dbh-CKO) mice’, and their control littermates’ electrocortical response in several contexts of enhanced arousal. Under enforced wakefulness (EW), or after cage change (CC), Hcrtr1Dbh-CKO mice exhibited a weakened ability to lower infra-θ frequencies (1–7 Hz), and mount a robust, narrow-bandwidth, high-frequency θ rhythm (~8.5 Hz). A fast-γ (55–80 Hz) response, whose dynamics closely parallelled θ, also diminished, while β/slow-γ activity (15–45 Hz) increased. Furthermore, EW-associated locomotion was lower. Surprisingly, nestbuilding-associated wakefulness, inversely, featured enhanced θ and fast-γ activities. Thus HCRT-to-NA signalling may fine-tune arousal, up in alarming conditions, and down during self-motivated, goal-driven behaviors. Lastly, slow-wave-sleep following EW and CC, but not nestbuilding, was severely deficient in slow-δ waves (0.75–2.25 Hz), suggesting that HCRT-to-NA signalling regulates the slow-δ rebound characterizing sleep after stress-associated arousal.
Hypocretin (orexin) is critical in sustaining theta/gamma-rich waking behaviors that drive sleep need.
Vassalli A, Franken P.
Proc Natl Acad Sci U S A. 2017 Jul 3;114(27):E5464-E5473. doi: 10.1073/pnas.1700983114. Epub 2017 Jun 19.PMID: 28630298
Summary
It is demonstrated that TDW instability causes profound blunting of EEG δ activity in subsequent slow-wave sleep, a measure gauging homeostatic sleep need, and proposed thatTDW identifies a discrete global brain activity mode that is regulated by context-dependent neuromodulators and acts as a major driver of sleep homeostasis.
Abstract
Significance As in narcolepsy patients, overall time spent awake and asleep is normal in mice lacking the wake-enhancing neuromodulator hypocretin/orexin. We discovered, however, that these mice, in baseline conditions, are impaired in maintaining theta-dominated wakefulness (TDW), a waking substate characteristic of goal-driven, explorative behaviors and associated with heightened θ/fast-γ activity. We demonstrate that TDW instability causes profound blunting of EEG δ activity in subsequent slow-wave sleep, a measure gauging homeostatic sleep need. In contrast, manually enforced waking induced unimpaired TDW expression and normal δ activity in recovery sleep. This suggests that TDW, not overall waking, drives sleep need, a hypothesis we verified by modeling the homeostatic process. We propose that Hcrt is critical for spontaneous waking but that enforced waking relies on other neuromodulators, such as norepinephrine. Hcrt gene inactivation in mice leads to behavioral state instability, abnormal transitions to paradoxical sleep, and cataplexy, hallmarks of narcolepsy. Sleep homeostasis is, however, considered unimpaired in patients and narcoleptic mice. We find that whereas Hcrtko/ko mice respond to 6-h sleep deprivation (SD) with a slow-wave sleep (SWS) EEG δ (1.0 to 4.0 Hz) power rebound like WT littermates, spontaneous waking fails to induce a δ power reflecting prior waking duration. This correlates with impaired θ (6.0 to 9.5 Hz) and fast-γ (55 to 80 Hz) activity in prior waking. We algorithmically identify a theta-dominated wakefulness (TDW) substate underlying motivated behaviors and typically preceding cataplexy in Hcrtko/ko mice. Hcrtko/ko mice fully implement TDW when waking is enforced, but spontaneous TDW episode duration is greatly reduced. A reformulation of the classic sleep homeostasis model, where homeostatic pressure rises exclusively in TDW rather than all waking, predicts δ power dynamics both in Hcrtko/ko and WT mouse baseline and recovery SWS. The low homeostatic impact of Hcrtko/ko mouse spontaneous waking correlates with decreased cortical expression of neuronal activity-related genes (notably Bdnf, Egr1/Zif268, and Per2). Thus, spontaneous TDW stability relies on Hcrt to sustain θ/fast-γ network activity and associated plasticity, whereas other arousal circuits sustain TDW during SD. We propose that TDW identifies a discrete global brain activity mode that is regulated by context-dependent neuromodulators and acts as a major driver of sleep homeostasis. Hcrt loss in Hcrtko/ko mice causes impaired TDW maintenance in baseline wake and blunted δ power in SWS, reproducing, respectively, narcolepsy excessive daytime sleepiness and poor sleep quality.
Hypocretin (orexin) biology and the pathophysiology of narcolepsy with cataplexy.
Liblau RS, Vassalli A, Seifinejad A, Tafti M.
Lancet Neurol. 2015 Mar;14(3):318-28. doi: 10.1016/S1474-4422(14)70218-2. Epub 2015 Feb 16.PMID: 25728441
Abstract
The discovery of hypocretins (orexins) and their causal implication in narcolepsy is the most important advance in sleep research and sleep medicine since the discovery of rapid eye movement sleep. Narcolepsy with cataplexy is caused by hypocretin deficiency owing to destruction of most of the hypocretin-producing neurons in the hypothalamus. Ablation of hypocretin or hypocretin receptors also leads to narcolepsy phenotypes in animal models. Although the exact mechanism of hypocretin deficiency is unknown, evidence from the past 20 years strongly favours an immune-mediated or autoimmune attack, targeting specifically hypocretin neurons in genetically predisposed individuals. These neurons form an extensive network of projections throughout the brain and show activity linked to motivational behaviours. The hypothesis that a targeted immune-mediated or autoimmune attack causes the specific degeneration of hypocretin neurons arose mainly through the discovery of genetic associations, first with the HLA-DQB1*06:02 allele and then with the T-cell receptor α locus. Guided by these genetic findings and now awaiting experimental testing are models of the possible immune mechanisms by which a specific and localised brain cell population could become targeted by T-cell subsets. Great hopes for the identification of new targets for therapeutic intervention in narcolepsy also reside in the development of patient-derived induced pluripotent stem cell systems.
Electroencephalogram paroxysmal θ characterizes cataplexy in mice and children.
Vassalli A, Dellepiane JM, Emmenegger Y, Jimenez S, Vandi S, Plazzi G, Franken P, Tafti M.
Brain. 2013 May;136(Pt 5):1592-608. doi: 10.1093/brain/awt069.PMID: 23616586
Summary
It is shown by deep recordings in mice that the cataplexy-associated state and hypersynchronous paroxysmal theta activities are highly enriched during catAPlexy in hypocretin/orexin knockout mice, contradict the hypothesis that catapLexy is a state similar to paradoxical sleep, even if long catap Lexies may evolve into paradoxicalSleep.
Abstract
Astute control of brain activity states is critical for adaptive behaviours and survival. In mammals and birds, electroencephalographic recordings reveal alternating states of wakefulness, slow wave sleep and paradoxical sleep (or rapid eye movement sleep). This control is profoundly impaired in narcolepsy with cataplexy, a disease resulting from the loss of orexin/hypocretin neurotransmitter signalling in the brain. Narcolepsy with cataplexy is characterized by irresistible bouts of sleep during the day, sleep fragmentation during the night and episodes of cataplexy, a sudden loss of muscle tone while awake and experiencing emotions. The neural mechanisms underlying cataplexy are unknown, but commonly thought to involve those of rapid eye movement-sleep atonia, and cataplexy typically is considered as a rapid eye movement sleep disorder. Here we reassess cataplexy in hypocretin (Hcrt, also known as orexin) gene knockout mice. Using a novel video/electroencephalogram double-blind scoring method, we show that cataplexy is not a state per se, as believed previously, but a dynamic, multi-phased process involving a reproducible progression of states. A knockout-specific state and a stereotypical paroxysmal event were introduced to account for signals and electroencephalogram spectral characteristics not seen in wild-type littermates. Cataplexy almost invariably started with a brief phase of wake-like electroencephalogram, followed by a phase featuring high-amplitude irregular theta oscillations, defining an activity profile distinct from paradoxical sleep, referred to as cataplexy-associated state and in the course of which 1.5-2 s high-amplitude, highly regular, hypersynchronous paroxysmal theta bursts (∼7 Hz) occurred. In contrast to cataplexy onset, exit from cataplexy did not show a predictable sequence of activities. Altogether, these data contradict the hypothesis that cataplexy is a state similar to paradoxical sleep, even if long cataplexies may evolve into paradoxical sleep. Although not exclusive to overt cataplexy, cataplexy-associated state and hypersynchronous paroxysmal theta activities are highly enriched during cataplexy in hypocretin/orexin knockout mice. Their occurrence in an independent narcolepsy mouse model, the orexin/ataxin 3 transgenic mouse, undergoing loss of orexin neurons, was confirmed. Importantly, we document for the first time similar paroxysmal theta hypersynchronies (∼4 Hz) during cataplexy in narcoleptic children. Lastly, we show by deep recordings in mice that the cataplexy-associated state and hypersynchronous paroxysmal theta activities are independent of hippocampal theta and involve the frontal cortex. Cataplexy hypersynchronous paroxysmal theta bursts may represent medial prefrontal activity, associated in humans and rodents with reward-driven motor impulse, planning and conflict monitoring.
Promoter architecture of mouse olfactory receptor genes.
Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, Vassalli A, Lazarevic D, Severin J, Vlachouli C, Simone R, Faulkner GJ, Kawai J, Daub CO, Zucchelli S, Hayashizaki Y, Mombaerts P, Lenhard B, Gustincich S, Carninci P.
Genome Res. 2012 Mar;22(3):486-97. doi: 10.1101/gr.126201.111. Epub 2011 Dec 22.PMID: 22194471
Summary
It is shown that a short genomic fragment flanking the major TSS of the OR gene Olfr160 (M72) can drive OSN-specific expression in transgenic mice.
Abstract
Odorous chemicals are detected by the mouse main olfactory epithelium (MOE) by about 1100 types of olfactory receptors (OR) expressed by olfactory sensory neurons (OSNs). Each mature OSN is thought to express only one allele of a single OR gene. Major impediments to understand the transcriptional control of OR gene expression are the lack of a proper characterization of OR transcription start sites (TSSs) and promoters, and of regulatory transcripts at OR loci. We have applied the nanoCAGE technology to profile the transcriptome and the active promoters in the MOE. nanoCAGE analysis revealed the map and architecture of promoters for 87.5% of the mouse OR genes, as well as the expression of many novel noncoding RNAs including antisense transcripts. We identified candidate transcription factors for OR gene expression and among them confirmed by chromatin immunoprecipitation the binding of TBP, EBF1 (OLF1), and MEF2A to OR promoters. Finally, we showed that a short genomic fragment flanking the major TSS of the OR gene Olfr160 (M72) can drive OSN-specific expression in transgenic mice.
Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice.
Vassalli A, Feinstein P, Mombaerts P.
Mol Cell Neurosci. 2011 Feb;46(2):381-96. doi: 10.1016/j.mcn.2010.11.001. Epub 2010 Nov 26.PMID: 21111823
Abstract
Odorant receptor (OR) genes constitute with 1200 members the largest gene family in the mouse genome. A mature olfactory sensory neuron (OSN) is thought to express just one OR gene, and from one allele. The cell bodies of OSNs that express a given OR gene display a mosaic pattern within a particular region of the main olfactory epithelium. The mechanisms and cis-acting DNA elements that regulate the expression of one OR gene per OSN – OR gene choice – remain poorly understood. Here, we describe a reporter assay to identify minimal promoters for OR genes in transgenic mice, which are produced by the conventional method of pronuclear injection of DNA. The promoter transgenes are devoid of an OR coding sequence, and instead drive expression of the axonal marker tau-β-galactosidase. For four mouse OR genes (M71, M72, MOR23, and P3) and one human OR gene (hM72), a mosaic, OSN-specific pattern of reporter expression can be obtained in transgenic mice with contiguous DNA segments of only ~300 bp that are centered around the transcription start site (TSS). The ~150bp region upstream of the TSS contains three conserved sequence motifs, including homeodomain (HD) binding sites. Such HD binding sites are also present in the H and P elements, DNA sequences that are known to strongly influence OR gene expression. When a 19mer encompassing a HD binding site from the P element is multimerized nine times and added upstream of a MOR23 minigene that contains the MOR23 coding region, we observe a dramatic increase in the number of transgene-expressing founders and lines and in the number of labeled OSNs. By contrast, a nine times multimerized 19mer with a mutant HD binding site does not have these effects. We hypothesize that HD binding sites in the H and P elements and in OR promoters modulate the probability of OR gene choice.
Sleep function: current questions and new approaches.
Vassalli A, Dijk DJ.
Eur J Neurosci. 2009 May;29(9):1830-41. doi: 10.1111/j.1460-9568.2009.06767.x. Epub 2009 Apr 28.PMID: 19473236
Comment in: Neuroscience: Sleepy and dreamless mutant mice. Dijk DJ, Winsky-Sommerer R.
Nature. 2016 Nov 17;539(7629):364-365. doi: 10.1038/nature20471. Epub 2016 Nov 2.PMID: 27806380
Summary
Wake experience‐induced local network changes may be sensed by the sleep homeostatic process and used to mediate sleep‐dependent events, benefiting network stabilization and memory consolidation.
Abstract
The mammalian brain oscillates through three distinct global activity states: wakefulness, non-rapid eye movement (NREM) sleep and REM sleep. The regulation and function of these ‘vigilance’ or ‘behavioural’ states can be investigated over a broad range of temporal and spatial scales and at different levels of functional organization, i.e. from gene expression to memory, in single neurons, cortical columns or the whole brain and organism. We summarize some basic questions that have arisen from recent approaches in the quest for the functions of sleep. Whereas traditionally sleep was viewed to be regulated through top-down control mechanisms, recent approaches have emphasized that sleep is emerging locally and regulated in a use-dependent (homeostatic) manner. Traditional markers of sleep homeostasis, such as the electroencephalogram slow-wave activity, have been linked to changes in connectivity and plasticity in local neuronal networks. Thus waking experience-induced local network changes may be sensed by the sleep homeostatic process and used to mediate sleep-dependent events, benefiting network stabilization and memory consolidation. Although many questions remain unanswered, the available data suggest that sleep function will best be understood by an analysis which integrates sleep’s many functional levels with its local homeostatic regulation.
Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse.
Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, Mombaerts P.
Neuron. 2009 Jan 29;61(2):220-33. doi: 10.1016/j.neuron.2008.11.010.PMID: 19186165
Abstract
The repertoire of approximately 1200 odorant receptors (ORs) is mapped onto the array of approximately 1800 glomeruli in the mouse olfactory bulb (OB). The spatial organization of this array is influenced by the ORs. Here we show that glomerular mapping to broad domains in the dorsal OB is determined by two types of olfactory sensory neurons (OSNs), which reside in the dorsal olfactory epithelium. The OSN types express either class I or class II OR genes. Axons from the two OSN types segregate already within the olfactory nerve and form distinct domains of glomeruli in the OB. These class-specific anatomical domains correlate with known functional odorant response domains. However, axonal segregation and domain formation are not determined by the class of the expressed OR protein. Thus, the two OSN types are determinants of axonal wiring, operate at a higher level than ORs, and contribute to the functional organization of the glomerular array.
Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice.
Grosmaitre X, Vassalli A, Mombaerts P, Shepherd GM, Ma M.
Proc Natl Acad Sci U S A. 2006 Feb 7;103(6):1970-5. doi: 10.1073/pnas.0508491103. Epub 2006 Jan 30. PMID: 16446455; PMCID: PMC1413638.
Abstract
A glomerulus in the mammalian olfactory bulb receives axonal inputs from olfactory sensory neurons (OSNs) that express the same odorant receptor (OR). Glomeruli are generally thought to represent functional units of olfactory coding, but there are no data on the electrophysiological properties of OSNs that express the same endogenous OR. Here, using patch clamp recordings in an intact epithelial preparation, we directly measured the transduction currents and receptor potentials from the dendritic knobs of mouse OSNs that express the odorant receptor MOR23 along with the green fluorescent protein. All of the 53 cells examined responded to lyral, a known ligand for MOR23. There were profound differences in response kinetics, particularly in the deactivation phase. The cells were very sensitive to lyral, with some cells responding to as little as 10 nM. The dynamic range was unexpectedly broad, with threshold and saturation in individual cells often covering three log units of lyral concentration. The potential causes and biological significance of this cellular heterogeneity are discussed. Patch clamp recording from OSNs that express a defined OR provides a powerful approach to investigate the sensory inputs to individual glomeruli.
Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor.
Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P.
Cell. 2004 Jun 11;117(6):833-46. doi: 10.1016/j.cell.2004.05.013. PMID: 15186782.
Abstract
Odorant receptors (ORs) provide the core determinant of identity for axons of olfactory sensory neurons (OSNs) to coalesce into glomeruli in the olfactory bulb. Here, using gene targeting in mice, we examine how the OR protein determines axonal identity. An OR::GFP fusion protein is present in axons, consistent with a direct function of ORs in axon guidance. When the OR coding region is deleted, we observe OSNs that coexpress other ORs that function in odorant reception and axonal identity. It remains unclear if such coexpression is normally prevented by negative feedback on OR gene choice. A drastic reduction in OR protein level produces axonal coalescence into novel, remote glomeruli. By contrast, chimeric ORs and ORs with minor mutations perturb axon outgrowth. Strikingly, the beta2 adrenergic receptor can substitute for an OR in glomerular formation when expressed from an OR locus. Thus, ORs have not evolved a unique function in axon guidance.
Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb.
Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P.
Neuron. 2002 Aug 15;35(4):681-96. doi: 10.1016/s0896-6273(02)00793-6. PMID: 12194868.
Abstract
An olfactory sensory neuron (OSN) expresses selectively one member from a repertoire of approximately 1000 odorant receptor (OR) genes and projects its axon to a specific glomerulus in the olfactory bulb. Both processes are here recapitulated by MOR23 and M71 OR minigenes, introduced into mice. Minigenes of 9 kb and as short as 2.2 kb are selectively expressed by neurons that do not coexpress the endogenous gene but coproject their axons to the same glomeruli. Deletion of a 395 bp upstream region in the MOR23 minigene abolishes expression. In this region we recognize sequence motifs conserved in many OR genes. Transgenic lines expressing the OR in ectopic epithelial zones form ectopic glomeruli, which also receive input from OSNs expressing the cognate endogenous receptor. This suggests a recruitment through homotypic interactions between OSNs expressing the same OR.