Phage-host interactions in mycolated bacteria
We investigate how phages engage with mycolated bacteria at the molecular level. Using cryo-electron tomography (cryo-ET) alongside genetics, biophysics and molecular modeling, we uncover how protein–protein interactions evolve and shape the host cell cycle. Our work focuses on the structural and molecular mechanisms of phage infection in environmental isolates from Lake Léman.
Molecular mechanisms of bacterial morphogenesis

Navarro, Vettiger et al., 2022
Bacterial cells display a variety of division and growth modes. We are interested in understanding the molecular mechanisms governing bacterial morphogenesis. To do that we apply an integrative approach combining our star technique cryo-electron tomography with bacterial genetics, molecular biology methods, computational approaches and other integrative imaging techniques.
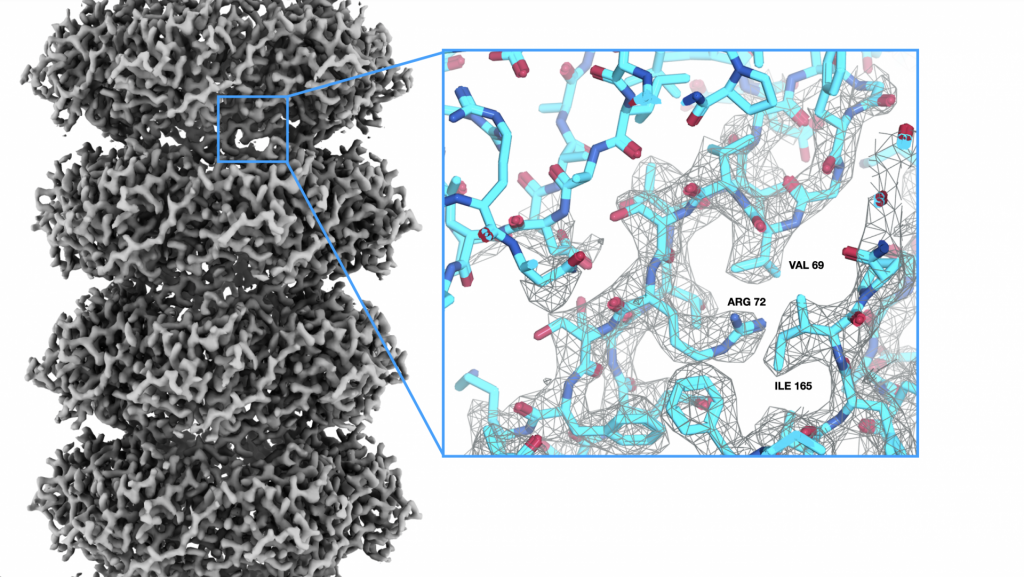
Cryo-electron tomography
Understanding of the 3D organization of cellular structure and biomolecules in situ is key to decipher their function in the cell. Cryo-electron tomography is a structural biology method that applied to living systems can determine the molecular architecture of cellular components and macromolecules in their native biological context. We apply and advance cryo-electron tomography technologies to image molecular landscapes for visual proteomics.

Image processing
We implement and develop computational tools for high-throughput cryo-electron tomographic data processing and subtomogram averaging. Our goal is to build computational strategies for quantitative analysis 3D volumes and in situ structure determination. To do that we develop cryoNAV, the software platform for high-throughput data handling of large-scale datasets, and contribute to the software package Dynamo for subtomogram averaging developed by Daniel Castaño’s lab.

