|
|

|
|
|
|
|
Facility News
Late spring is pretty harsh this year with pollen and allergies, not that easy to go outside without a lot of sneezing 🤧 and red eyes ! But you know what can help ? Being in our amazingly cocoon-y facility, performing top of the art flow cytometry ! Protect yourself a bit more until pollen is down and the summer is truly here !
|
|
In this month FACS Tips, we cover the basics of sorting. It's true that in our facility, you don't directly perform the sorts, the power rangers of flow do it for you. Yet, knowing how it works and what truly matter for it to succeed can help you prepare the sample in the best way possible !
|
|
Ionna Rota won last month quiz, congratulations to you !
|
|
|
Each month, we will give away one of those special and unique mug designed by the FCF team. Answer correctly to our 3 questions and you can have a chance to win !
Please take few minutes to answer the quiz HERE.
|
|
|
|
For the Epalinges site, a reminder :
|
On June 15th, we will perform a DIVA maintenance on the LSRIIa, LSRIIb and LSRII SORP. We will reinstall the software to clean the database and hopefully make it run a bit smoother.
Since the software will be reinstalled, you will lose your personal account and any data that is under your profile but not exported on the data drive. Everything exported on the D:/ Drive (FCS files and Experiment Templates) will remain and be saved.
As a consequence, please check that you have saved your data correctly before that date.
After that installation, we will create a profile per lab and each member will create a folder in its own name under that profile.
If you are not sure how to do it, please come see someone from the FCF Team to help you.
|
|
|
|
|
|
|
|
FACS Tips
|
Cell Sorting – Less (Cells) is More (Purity)
|
We ask a lot of cytometers. We need them to look at these millions to billions of events we’ve prepared, with potentially over 20 markers, and oh… we also want them to do it quite quickly. With sorters, we even ask a bit more, which is to say, lets also collect certain populations of interest out of those millions or billions of cells, without contaminating it with any of the other events that are passing through the machine at rates as high as 15,000 events per second. How sorters are able to separate individual cells out from such a rapid flow of heterogeneous sample is certainly impressive, but also understanding it better can allow us to better customize our sorting experiments to fit our end goals. Sorting is a balance between Purity, Yield, and Speed. Depending on your initial concentration, and how much you need of a desired population for your next steps, we can plan an approach to get there considering these 3 factors. For example, there are several sorting modes provided by the software to customize this approach, Yield, Purity, and Single Cell, and depending on what you want out of your experiment one may be better than others.
|
|
|
|
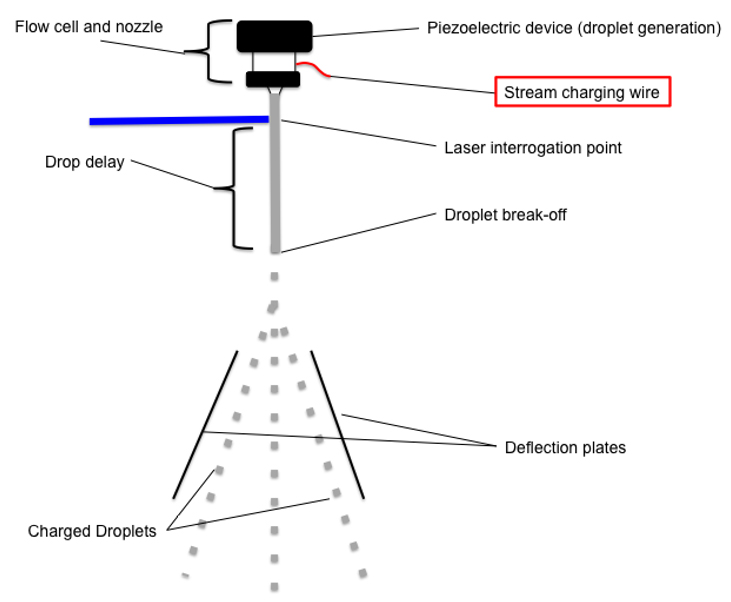
Image from https://expert.cheekyscientist.com/how-droplets-are-charged-drop-delays-determined-during-electrostatic-cell-sorting-experiment/
|
|
|
While the fluidics of a sorter are almost identical to what you would see on a typical analyzer, there’s a decent bit of extra equipment to make sorting possible. A key component of sorting is taking a solid stream of fluid, and breaking it apart into individual droplets, so they can be separately collected. This is achieved by injecting the stream into air. However, this will only occur in a stable and controlled manner if we set a stationary wave of vibration of known frequency and amplitude through the fluid stream. In a sorter, this vibration is produced by a transducer, which is generally a piezoelectric crystal acoustically coupled to the nozzle. The vibration causes the stream to break into drops at a specific, and ideally consistent point, called the Drop Break Off. An important calculation here, is the time between the interrogation point and the exact point where the first droplet breaks off from the stream, which is called the Drop delay. What this means is that when a cell of interest passes through the interrogation point, and its fluorescence is detected and measured, the machine can then predict when it will arrive at the drop breakoff point, and what drop it is likely to be in. This prediction is what leads into our various sort mode decisions.
|
|
|
|
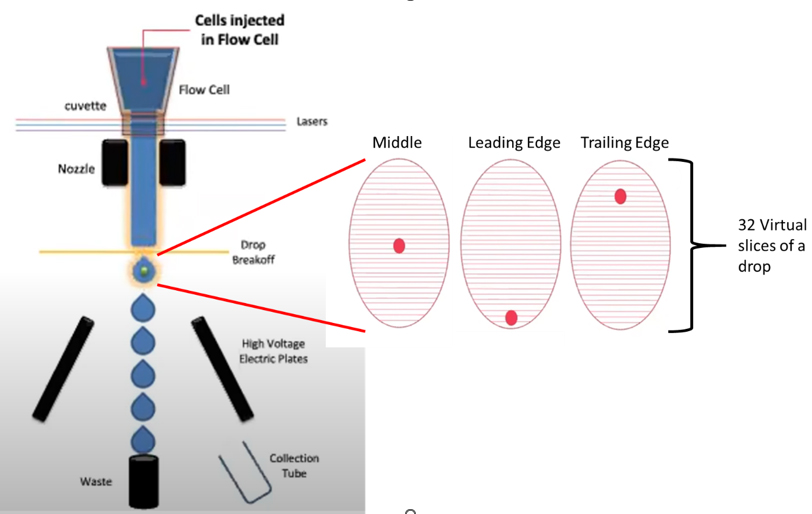
Image from OpenFlow Cytometry Sorting webinar https://www.youtube.com/watch?v=P6e9EbNdS64
|
|
|
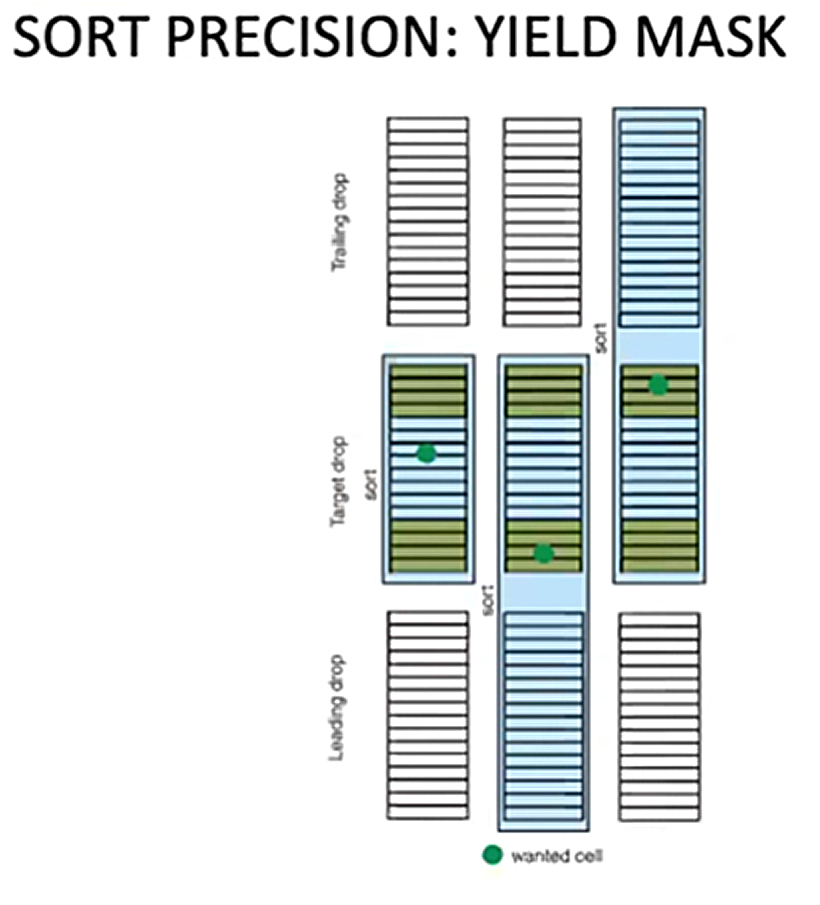
When our cell of interest passes through the interrogation point, it’s still in a stream, so we have to anticipate where it will be by the time it breaks off into the first drop. We know thanks to the drop delay the time it should take, but this means the cell could be in the middle of the drop, which is ideal, or other times at the leading or trailing edge. DIVA actually divides each drop into 32 virtual slices to make this estimation of its position in the drop. Cells can also deviate from their expected arrival at the drop breakoff, sometimes faster sometimes slower, meaning that if they were already anticipated at the leading or trailing edge of a drop they might jump one drop ahead or behind the initial calculation. If we only sort one drop, we risk losing these cells, but if sort two drops we can still get as much of the desired population as possible. There is some risk with sorting multiple drops though, as it can increase the odds we get something we don’t want, but it also will inflate our collection volume. This is where are different sorting modes come into play.
|
|
|
|
Image from OpenFlow Cytometry Sorting webinar https://www.youtube.com/watch?v=P6e9EbNdS64
|
|
Yield Mode
|
As its name suggests, sorting in yield mode will give you the highest possible return of cells of interest, even if that means occasionally giving you something you don’t want. Yield mode is most concerned with only the target drop to be sorted and the position of the event of interest within it. It will sort two drops if it’s unsure whether an event of interest has the potential of jumping one drop ahead or behind. It will also sort a cell of interest if there is a non-target event likely to come with it. We would recommend using yield mode in experiment were getting the largest number of cells is the most advantageous for you, also if you don’t mind having a larger final volume. The yield mode could also be used as a first-round sort for enrichment of target particles, followed by a sort for purity when samples are very large and concentrated.
|
|
|
|
Image from OpenFlow Cytometry Sorting webinar https://www.youtube.com/watch?v=P6e9EbNdS64
|
|
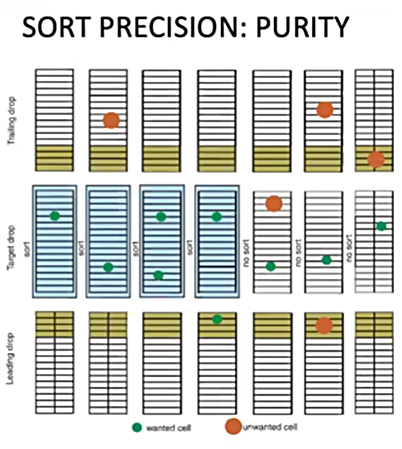
Purity Mode
|
In purity mode the sorter is much stricter. Purity mode takes into consideration the 32 slice mask of the drops above and below the target drop. If there is a chance that a non-targeted event has entered into the defined leading or tailing area of the drop before or after, then it will no longer sort your cell of interest for fear of contamination. This means you will lose the cell and have a smaller final number, but with better purity. This is the mode we use by default when sorting. It provides a nice combination of speed and purity, and hopefully won’t lose too many cells.
|
|
|
|
Image from OpenFlow Cytometry Sorting webinar https://www.youtube.com/watch?v=P6e9EbNdS64
|
|
|
|
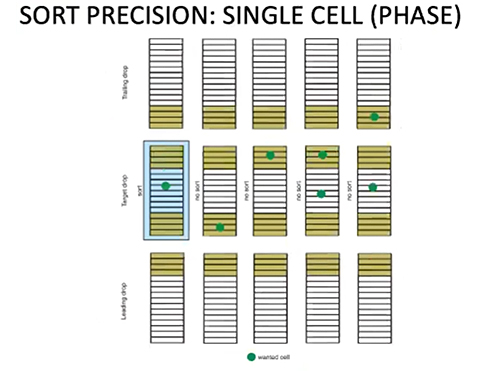
Single Cell Mode
This is the strictest mask. It is paying attention to both the 32 slices in the drop of interest and the 32 slices in the drop above and below. This will only ever sort one drop, and only under the condition that the cell of interest is predicted to be in the center of the drop. This mode will produce the greatest purity and will also give you the smallest possible final volume, but it will also take the longest and lose the most cells of interest to waste. This mode is most frequently used for plate sorting, but can also be valuable in other situations where highest purity is most important and strictest control of the final volume matters.
|
|
Lots of factors can influence our sort, and there are many variables we can think about when it comes to best preparing both the machine and our sample. For example, the nozzle we choose. With the 70um nozzle, which runs faster at a higher pressure and frequency, you get about 80,000 to 90,000 drops a second, while the 100um nozzle it will be only be 30,000 to 40,000 drops a second. The 70um nozzle drops will also be smaller, about 1nl vs 4nl for the 100um nozzle. However, we can only use the 70um nozzle for smaller cells, and the higher pressure could potential put more stress on the cells, which could be a factor if the cells are particularly vulnerable to begin with. By balancing our sort mode choice with the nozzle selected and the sample quality, along with other factors in machine setup, we can optimize best for each individual experiment. If you have any questions about your sort as always feel free to reach out to the staff at the FCF for advice.
|
|
|
|
|