|
|

|
|
|
|
|
Facility News
As Spring🌱is around the corner and nature comes back to life, we are also trying to bring improvements and renewal to our facility. As you may have notice, we asked for your feedback about the FCF, if you haven't had time to look into it, please consider doing it now to make sure we can help you better in the future ! You have until March 7th to do it HERE.
|
|
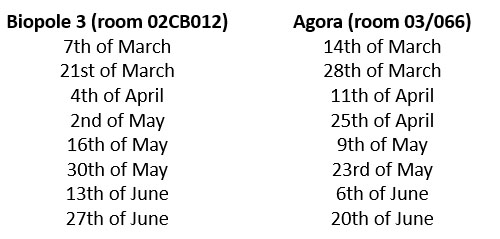
We've recently changed the way we do practical trainings, due to high demand. From now on, Jean-François Mayol will alternate in person practical training every tuesday morning from 10 am to 12 pm at the Biopole and from 9:30 am to 11:30 am at the Agora. The training is open to new users and current users who would like a refresher. Please contact him first to make sure there are spots left. You will find the dates for each location below :
|
|
|
In this month FACS Tips, we're explaining key steps to make sure your data is properly optimised for dimension reduction ! It doesn't have to be that complicated and we hope it will encourage you to try out some of the sofwares we mentioned in previous newsletters.
|
|
Mélanie Tichet won last month quiz, congratulations to you, you get this sadly very accurate mug 😉! You've asked us about the Toblerone bar not be awarded anymore. Would you like it to be back ? Voice your opinion and it might soon be back !
|
|

|
|
Each month, we will give away one of those special and unique mug designed by the FCF team so please take few minutes to answer the quiz HERE.
|
|
|
|
|
|
FACS Tips
|
Dimension Reduction Sample Prep Process
|
While dimension reduction in general, and more specifically tSNE, have been covered in previous newsletters, the process of preparing samples for analysis using dimension reduction methods can be confusing. As dimension reduction maps change from sample to sample, to get a reproducible comparison across several samples multiple files need to be concatenated together and limited in the number of events. Once you have multiple files concatenated together though, we still need to find a way to break them apart to look at them individually, requiring the need to add keywords. These steps are not routinely part of a conventional analysis, as we usually have recorded as much as needed and can compare everything separately. Also, to get the most out of our data, it’s best to perform a data cleaning to remove erroneous data. Learning these steps are not hard and will be necessary to achieve a proper dimension reduction analysis for whatever tool of interest you would like to use.
|
Quality Control Algorithms
|
There are a variety of tools available in FlowJo to detect quality issues, and can be a great starting point for preparing your samples for dimension reduction analysis. Three of the most well known are FlowAI, FlowClean, and PeacoQC. They all perform a similar function of running an automated check on your samples to remove abhorrent events caused by clogs, air bubbles, and any other abnormalities. The output is a presented as “Good Events” “Bad Events” subgroup that you can further gate from. In a future newsletter we will go into greater depths as to how each works and which one might be right for your experiment.
|
In longitudinal flow cytometry studies, one problem we have to overcome is batch effects. By combing files together across multiple timepoints we can have technical variations influence our results. FlowJo has added a plugin to address this problem, called CytoNorm, which can be applied to your samples as long as you have provided a similar control across timepoints, for example a positive control run in each batch of the experiment. While it’s too in-depth to go into here, the plugin applies FlowSom clustering on your shared controls across time and that allows for an adjustment to be made that corrects batch effect errors. The best approach for a control is to take a sample from one subject, preserve and freeze, and then use each time a new batch is performed. Unfortunately, FMO data has not shown much success, while single stain controls have some potential.
|
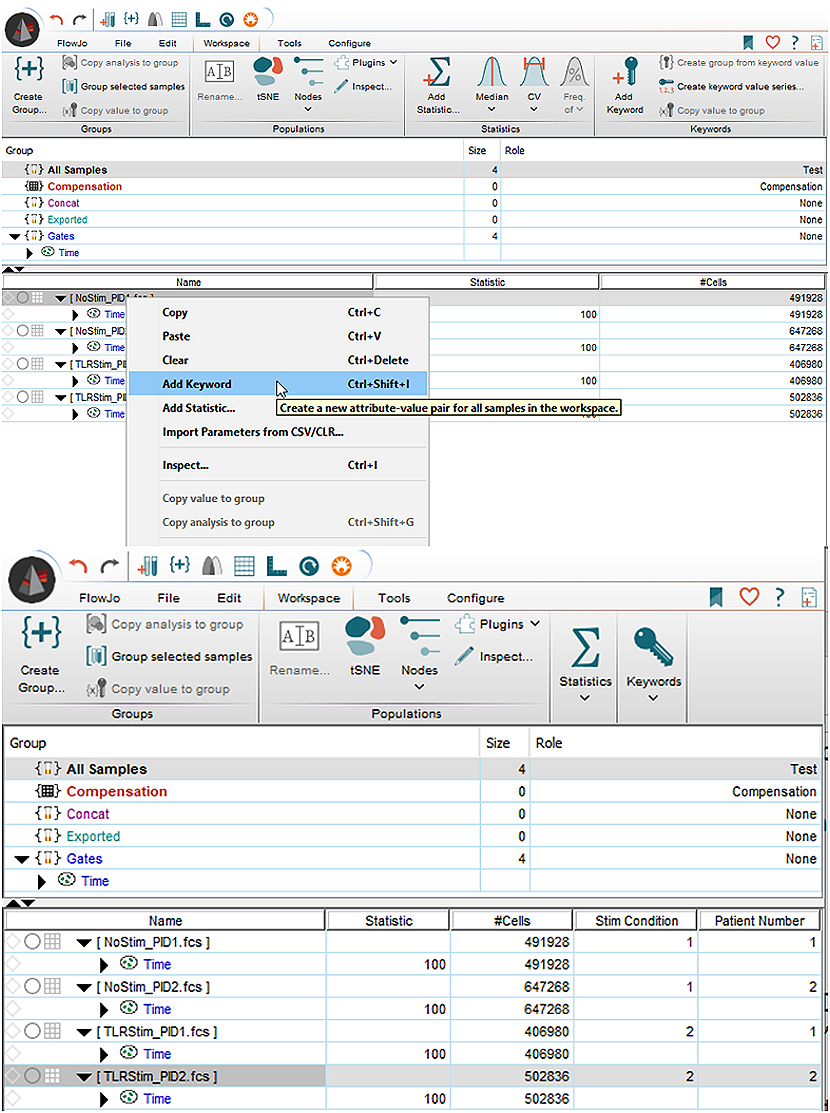
A new keyword will need to be added for each sample before concatenation, such that they can still be separated. This can be done by right clicking the sample and selecting “Add Keyword”. For this example, I’ve added two keywords, “Stim Condition” and “Patient Number”. Next, we assign a Keyword Value for each of these. For this, just click on the column and insert the desired text, for me here it’s 1, 1, 2, 2 and 1, 2, 1, 2. Keywords can also be added in DIVA before you record your files, if you know you plan on doing tSNE at the start of an experiment.
|
|
|
|
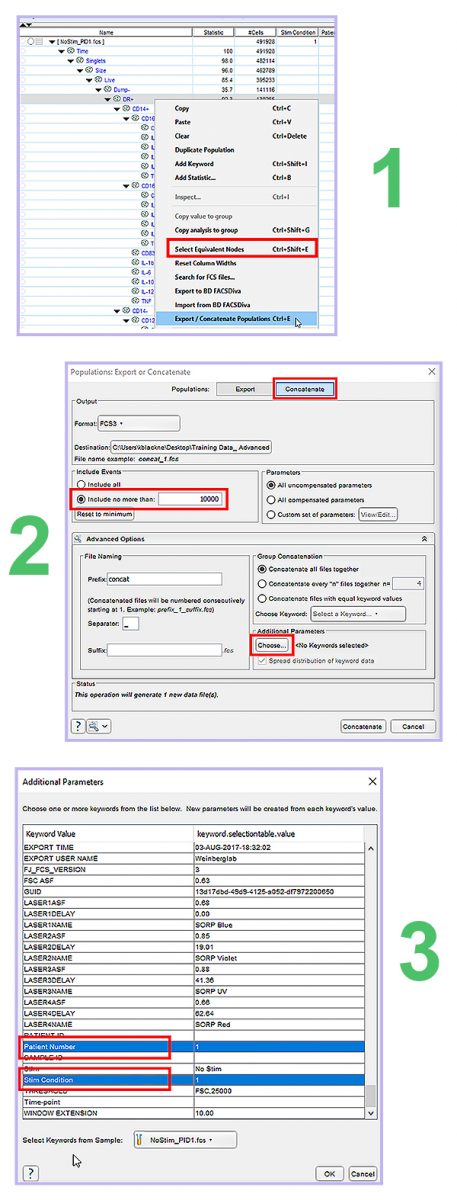
Concatenate is a feature in FlowJo that allows you to combine files together into one FCS file. To make the most of our future dimension reduction analysis, we will concatenate our samples further downstream from our usual FSCxSSC opening gate. It’s best to select a population with dead cells, doublets, and uninteresting populations already removed. To do this, we will right click on the parent population of interest in one sample and then press “Select Equivalent Nodes”, then right click again and press “Export/Concatenate Populations”. Select the concatenate tab on the top right. Because tSNE has limitations to the number of events it can process without crashing, we will limit it to include no more than 10,000 events per sample, and then we will add both the Patient Number and Stim Condition keywords we created. Once this is set up we can hit concatenate in the bottom right, and have this concatenated file selected to open in the existing workspace.
|
|
|
|
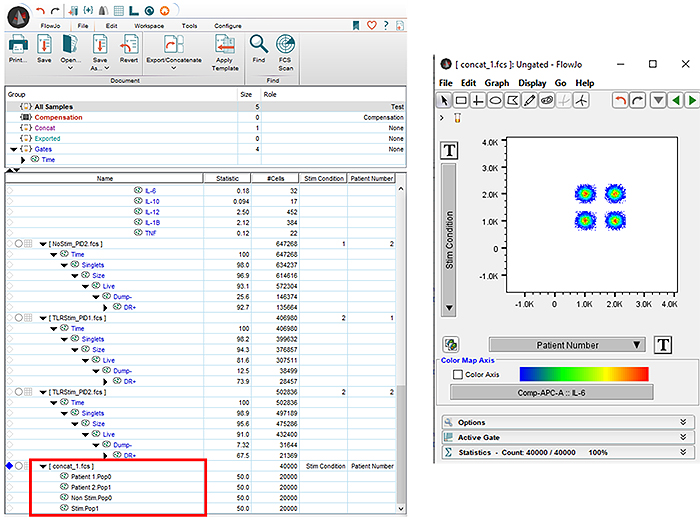
Now, by evaluating the concatenated file from your existing worksheet, you can set up a bivariate plot to see the separate Stimulation conditions as well as the patient numbers. From here you can gate it how you would like for the tSNE to be evaluated, either by Stimulation, Patient, or individually.
|
|
|
Run your Dimension Reduction
|
|
Lastly, we can now select the concatenated sample and run the dimension reduction of choice and apply it to our divided conditions/patients and compare.
|
|
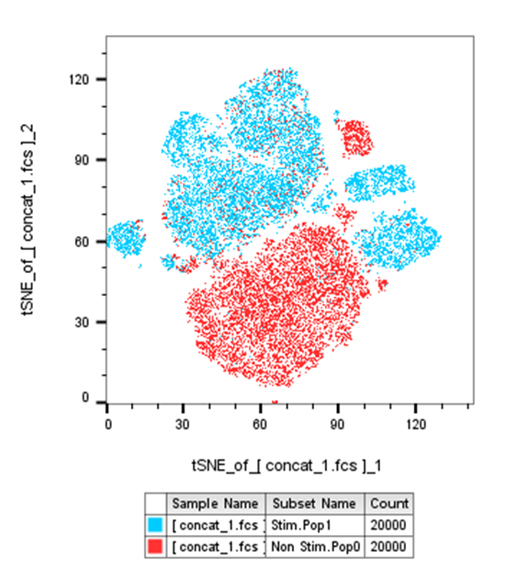
|
|
Dimension reduction algorithms provide exciting new opportunities for data analysis, however they come with added challenges in data preparation. Without properly organizing our data beforehand we can end up with misleading or confusing results. Even without performing a dimension reduction algorithm, the data cleaning pipeline is a valuable tool for our conventional hierarchical gating strategy. If you would like some sample data along with a more detailed step-by-step description to follow along on the process described here, including guides for tSNE and UMAP feel free to ask us here in the FCF and we can provide you with the files.
|
|
|
|
|
|
|