High-throughput biotechnologies are able to generate huge amounts of highly detailed molecular data from biological samples, including genomic, transcriptomic, proteomic and metabolomic data. These technologies are therefore supremely suited for studying the complexities of the microbial communities associated with bees. In theory, this allows us to obtain a global picture of the effects, population dynamics, interactions, exchange and evolution of these communities within and between bee species. To make this possible, sequence data produced by different laboratories need to be consistently analyzed and archived, allowing integration and subsequent exploitation by scientists from different research areas, such as bee pathology, microbial ecology, and evolution. This consortium was establisched through the project “BeeBiome: Omic approaches for understanding bee-microbe relationships”, a workshop organized in 2014 at the National Evolutionary Synthesis Center (NESCent) in Durham, USA supported by a NESCent Catalysis meeting grant, NSF #EF- 0905606, that led to the formation of the Beebiome consortium, representing over 30 scientists from Europe and the USA, bee-scientist, ecologists and bioinformaticians, working on bee-associated microbes. This group aims to:
- identify the most burning questions regarding bee microbiome interactions, evolution, biodiversity, microbial patterns and the effect of the environment on its composition,
- define an appropriate strategy to address these questions, including the design of tools such as a dedicated database,
- establish future collaborative efforts to fund the proposed research.
Schematic representation of the Consortium Aims: 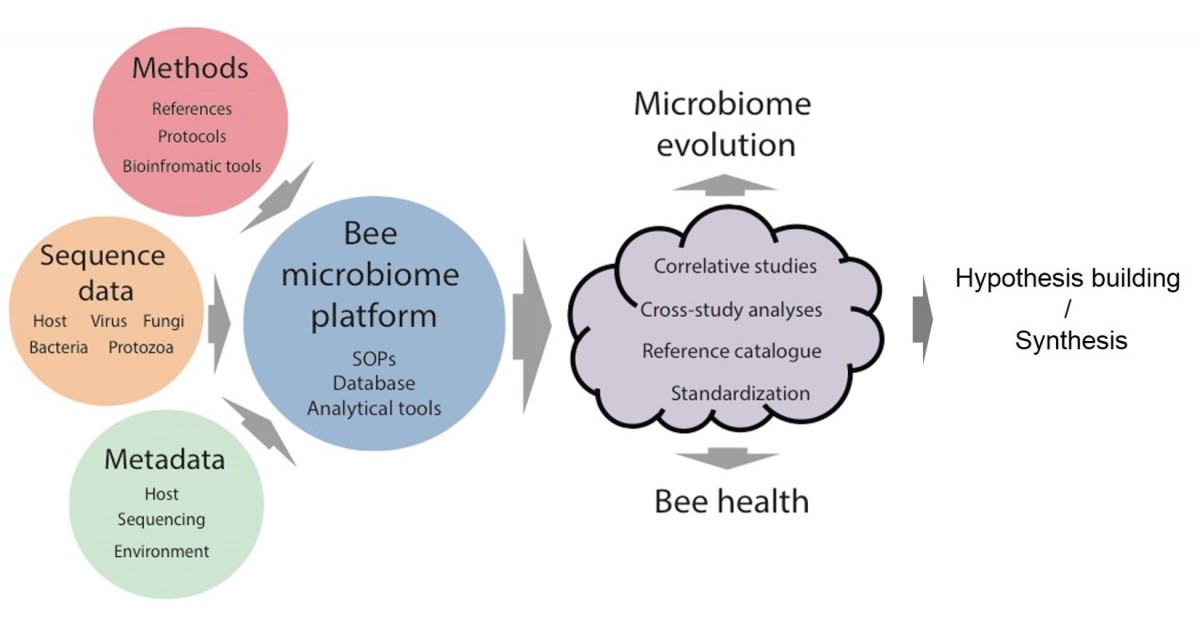
A primary aim of the Bee Microbiome Consortium is a centralized database and data analysis platform for bee microbiome studies. Such a platform is needed to systematically archive, curate and process the large amounts of data generated by laboratories worldwide; to make this data available to researchers in the field; to facilitate cross-study analyses, and to standardize analytical approaches to microbiome studies. Communal approaches to multiple, integrated data-sets maximize the use and value of such data, leading to more robust analyses and conclusions and to novel, research-driven, hypotheses about bee health, microbial ecology and microbiome evolution.
